| Full name: sodium channel and clathrin linker 1 | Alias Symbol: hCAP-1A|FLJ30655 | ||
| Type: protein-coding gene | Cytoband: 4q28.2 | ||
| Entrez ID: 132320 | HGNC ID: HGNC:26406 | Ensembl Gene: ENSG00000151466 | OMIM ID: 611399 |
| Drug and gene relationship at DGIdb | |||
Expression of SCLT1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | SCLT1 | 132320 | 4268 | -0.3092 | 0.0063 | |
| GSE53624 | SCLT1 | 132320 | 4268 | -0.1193 | 0.2201 | |
| GSE97050 | SCLT1 | 132320 | A_33_P3340404 | 0.0778 | 0.7212 | |
| SRP007169 | SCLT1 | 132320 | RNAseq | 0.1631 | 0.6941 | |
| SRP008496 | SCLT1 | 132320 | RNAseq | 0.3505 | 0.2028 | |
| SRP064894 | SCLT1 | 132320 | RNAseq | 0.3482 | 0.0418 | |
| SRP133303 | SCLT1 | 132320 | RNAseq | 0.3794 | 0.0248 | |
| SRP159526 | SCLT1 | 132320 | RNAseq | 0.1981 | 0.6029 | |
| SRP193095 | SCLT1 | 132320 | RNAseq | -0.0013 | 0.9895 | |
| SRP219564 | SCLT1 | 132320 | RNAseq | 0.1350 | 0.6647 | |
| TCGA | SCLT1 | 132320 | RNAseq | 0.2033 | 0.0145 |
Upregulated datasets: 0; Downregulated datasets: 0.
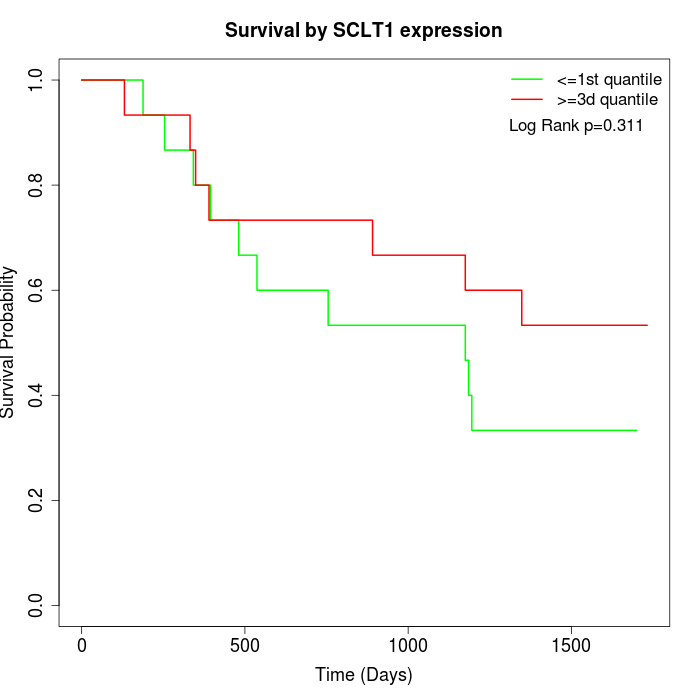
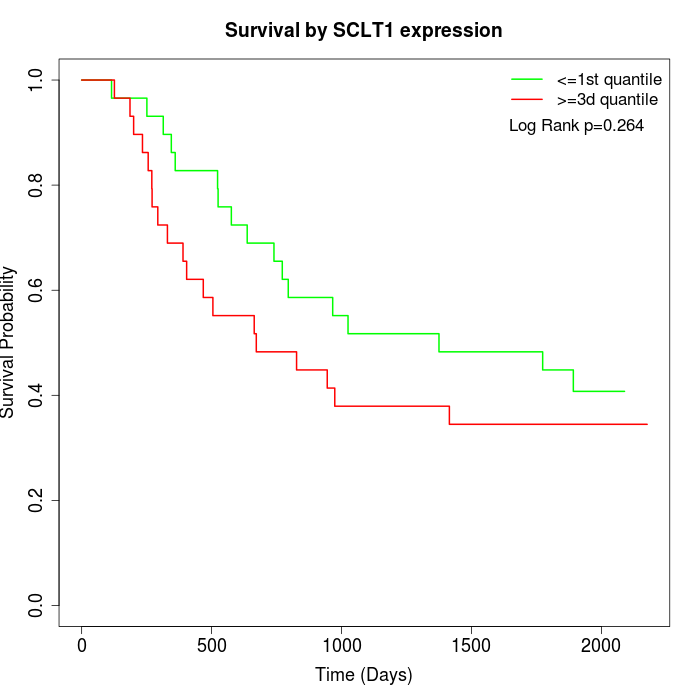
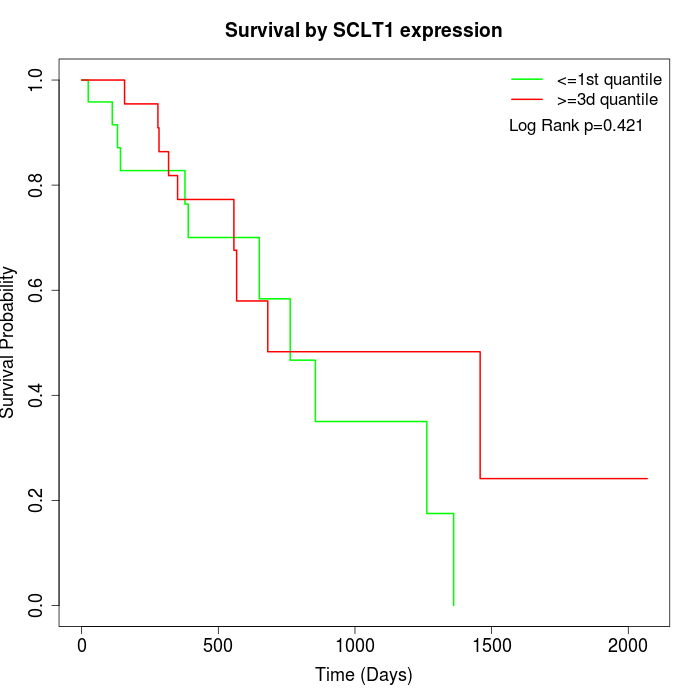
Survival by SCLT1 expression:
Note: Click image to view full size file.
Copy number change of SCLT1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SCLT1 | 132320 | 0 | 12 | 18 | |
| GSE20123 | SCLT1 | 132320 | 0 | 12 | 18 | |
| GSE43470 | SCLT1 | 132320 | 0 | 12 | 31 | |
| GSE46452 | SCLT1 | 132320 | 1 | 36 | 22 | |
| GSE47630 | SCLT1 | 132320 | 0 | 22 | 18 | |
| GSE54993 | SCLT1 | 132320 | 9 | 0 | 61 | |
| GSE54994 | SCLT1 | 132320 | 1 | 11 | 41 | |
| GSE60625 | SCLT1 | 132320 | 0 | 1 | 10 | |
| GSE74703 | SCLT1 | 132320 | 0 | 10 | 26 | |
| GSE74704 | SCLT1 | 132320 | 0 | 6 | 14 | |
| TCGA | SCLT1 | 132320 | 9 | 34 | 53 |
Total number of gains: 20; Total number of losses: 156; Total Number of normals: 312.
Somatic mutations of SCLT1:
Generating mutation plots.
Highly correlated genes for SCLT1:
Showing top 20/28 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SCLT1 | METTL14 | 0.720307 | 3 | 0 | 3 |
| SCLT1 | SYNJ1 | 0.699106 | 3 | 0 | 3 |
| SCLT1 | CPVL | 0.698091 | 3 | 0 | 3 |
| SCLT1 | GABPA | 0.69569 | 3 | 0 | 3 |
| SCLT1 | RGS18 | 0.688938 | 3 | 0 | 3 |
| SCLT1 | CREBL2 | 0.680691 | 3 | 0 | 3 |
| SCLT1 | P2RY13 | 0.672346 | 3 | 0 | 3 |
| SCLT1 | CD226 | 0.670772 | 3 | 0 | 3 |
| SCLT1 | GIMAP5 | 0.670187 | 3 | 0 | 3 |
| SCLT1 | CST7 | 0.667111 | 3 | 0 | 3 |
| SCLT1 | CEP44 | 0.662467 | 3 | 0 | 3 |
| SCLT1 | DPYD | 0.660098 | 3 | 0 | 3 |
| SCLT1 | POLK | 0.656432 | 3 | 0 | 3 |
| SCLT1 | FLI1 | 0.655707 | 3 | 0 | 3 |
| SCLT1 | TNFAIP8L2 | 0.654352 | 3 | 0 | 3 |
| SCLT1 | PWWP2A | 0.651336 | 3 | 0 | 3 |
| SCLT1 | PLCB2 | 0.649209 | 3 | 0 | 3 |
| SCLT1 | TLR3 | 0.643902 | 3 | 0 | 3 |
| SCLT1 | CEP120 | 0.643414 | 3 | 0 | 3 |
| SCLT1 | ZNF548 | 0.636169 | 3 | 0 | 3 |
For details and further investigation, click here