| Full name: cAMP responsive element binding protein like 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 12p13.1 | ||
| Entrez ID: 1389 | HGNC ID: HGNC:2350 | Ensembl Gene: ENSG00000111269 | OMIM ID: 603476 |
| Drug and gene relationship at DGIdb | |||
Expression of CREBL2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CREBL2 | 1389 | 201989_s_at | -0.3987 | 0.4612 | |
| GSE20347 | CREBL2 | 1389 | 201989_s_at | -0.1267 | 0.4658 | |
| GSE23400 | CREBL2 | 1389 | 201989_s_at | 0.1446 | 0.1099 | |
| GSE26886 | CREBL2 | 1389 | 201989_s_at | 0.0710 | 0.8113 | |
| GSE29001 | CREBL2 | 1389 | 201989_s_at | 0.4440 | 0.1564 | |
| GSE38129 | CREBL2 | 1389 | 201989_s_at | -0.2456 | 0.1360 | |
| GSE45670 | CREBL2 | 1389 | 201989_s_at | -0.2861 | 0.0689 | |
| GSE53622 | CREBL2 | 1389 | 61953 | -0.3544 | 0.0003 | |
| GSE53624 | CREBL2 | 1389 | 61953 | -0.3480 | 0.0032 | |
| GSE63941 | CREBL2 | 1389 | 201989_s_at | -0.8322 | 0.0080 | |
| GSE77861 | CREBL2 | 1389 | 201989_s_at | 0.3799 | 0.2903 | |
| GSE97050 | CREBL2 | 1389 | A_24_P56194 | -0.0284 | 0.9387 | |
| SRP007169 | CREBL2 | 1389 | RNAseq | 0.5097 | 0.2443 | |
| SRP008496 | CREBL2 | 1389 | RNAseq | 0.7253 | 0.0095 | |
| SRP064894 | CREBL2 | 1389 | RNAseq | 0.0899 | 0.6748 | |
| SRP133303 | CREBL2 | 1389 | RNAseq | 0.2976 | 0.1526 | |
| SRP159526 | CREBL2 | 1389 | RNAseq | -0.0469 | 0.8974 | |
| SRP193095 | CREBL2 | 1389 | RNAseq | -0.1736 | 0.1238 | |
| SRP219564 | CREBL2 | 1389 | RNAseq | 0.0408 | 0.9031 | |
| TCGA | CREBL2 | 1389 | RNAseq | -0.1880 | 0.0002 |
Upregulated datasets: 0; Downregulated datasets: 0.
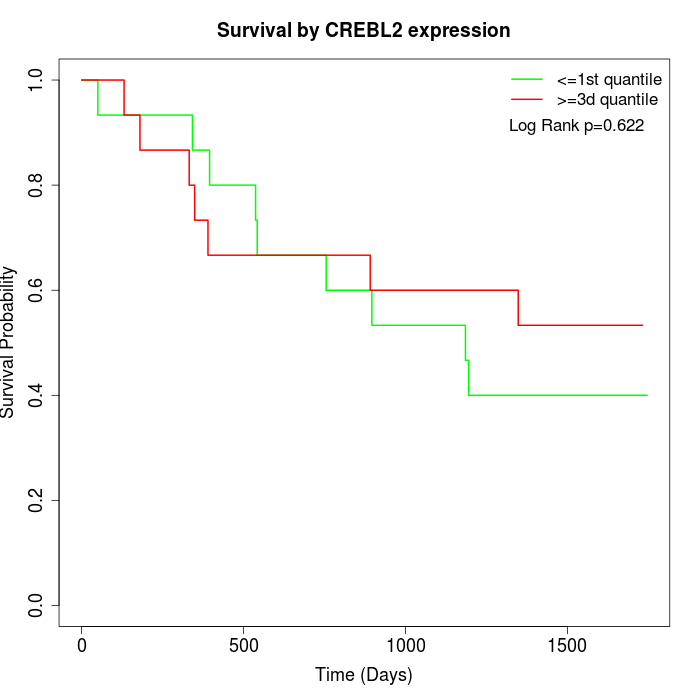
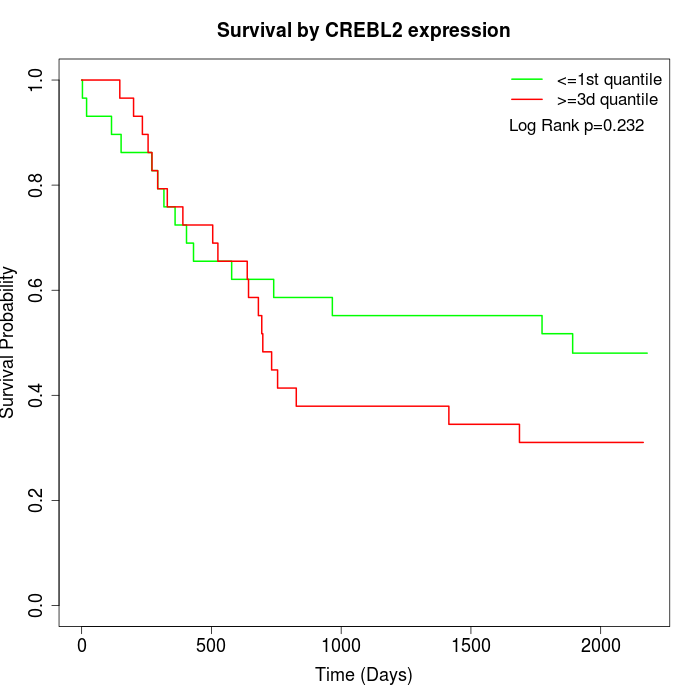
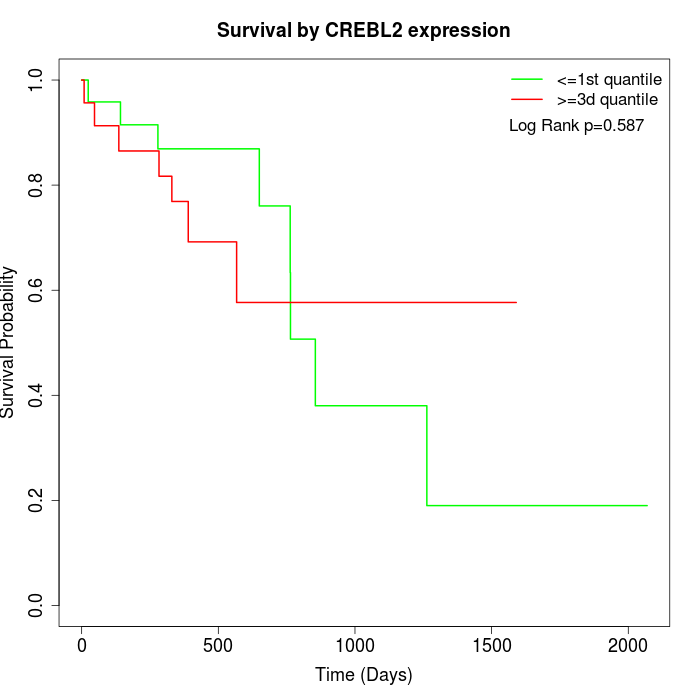
Survival by CREBL2 expression:
Note: Click image to view full size file.
Copy number change of CREBL2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CREBL2 | 1389 | 6 | 2 | 22 | |
| GSE20123 | CREBL2 | 1389 | 6 | 2 | 22 | |
| GSE43470 | CREBL2 | 1389 | 10 | 4 | 29 | |
| GSE46452 | CREBL2 | 1389 | 10 | 1 | 48 | |
| GSE47630 | CREBL2 | 1389 | 13 | 2 | 25 | |
| GSE54993 | CREBL2 | 1389 | 2 | 9 | 59 | |
| GSE54994 | CREBL2 | 1389 | 10 | 2 | 41 | |
| GSE60625 | CREBL2 | 1389 | 0 | 1 | 10 | |
| GSE74703 | CREBL2 | 1389 | 9 | 3 | 24 | |
| GSE74704 | CREBL2 | 1389 | 4 | 1 | 15 | |
| TCGA | CREBL2 | 1389 | 39 | 7 | 50 |
Total number of gains: 109; Total number of losses: 34; Total Number of normals: 345.
Somatic mutations of CREBL2:
Generating mutation plots.
Highly correlated genes for CREBL2:
Showing top 20/903 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CREBL2 | GIMAP5 | 0.806329 | 3 | 0 | 3 |
| CREBL2 | TM2D3 | 0.736923 | 4 | 0 | 4 |
| CREBL2 | CXCL5 | 0.724334 | 3 | 0 | 3 |
| CREBL2 | TXNDC11 | 0.724329 | 3 | 0 | 3 |
| CREBL2 | AEBP2 | 0.720508 | 4 | 0 | 3 |
| CREBL2 | GSTA3 | 0.716625 | 3 | 0 | 3 |
| CREBL2 | ACSL5 | 0.714962 | 3 | 0 | 3 |
| CREBL2 | GIMAP1 | 0.707206 | 6 | 0 | 5 |
| CREBL2 | KLRG1 | 0.702979 | 4 | 0 | 4 |
| CREBL2 | NEK3 | 0.701996 | 3 | 0 | 3 |
| CREBL2 | CYYR1 | 0.698532 | 5 | 0 | 5 |
| CREBL2 | TNFSF12 | 0.693332 | 3 | 0 | 3 |
| CREBL2 | AP3S2 | 0.687137 | 3 | 0 | 3 |
| CREBL2 | COQ10B | 0.684685 | 4 | 0 | 4 |
| CREBL2 | MIER1 | 0.684542 | 3 | 0 | 3 |
| CREBL2 | DNAJB6 | 0.683803 | 3 | 0 | 3 |
| CREBL2 | SATB1 | 0.683265 | 3 | 0 | 3 |
| CREBL2 | PEAK1 | 0.682265 | 5 | 0 | 5 |
| CREBL2 | LYVE1 | 0.681785 | 6 | 0 | 6 |
| CREBL2 | FCGRT | 0.68092 | 8 | 0 | 6 |
For details and further investigation, click here