| Full name: smooth muscle and endothelial cell enriched migration/differentiation-associated lncRNA | Alias Symbol: | ||
| Type: non-coding RNA | Cytoband: 11q24.3 | ||
| Entrez ID: 100507392 | HGNC ID: HGNC:44177 | Ensembl Gene: ENSG00000254703 | OMIM ID: 615815 |
| Drug and gene relationship at DGIdb | |||
Expression of SENCR:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SENCR | 100507392 | 239641_at | -0.0187 | 0.9576 | |
| GSE26886 | SENCR | 100507392 | 239641_at | -0.0758 | 0.6960 | |
| GSE45670 | SENCR | 100507392 | 239641_at | -0.1603 | 0.1466 | |
| GSE53622 | SENCR | 100507392 | 80283 | -0.1321 | 0.0194 | |
| GSE53624 | SENCR | 100507392 | 80283 | -0.0258 | 0.7931 | |
| GSE63941 | SENCR | 100507392 | 239641_at | 0.1155 | 0.7760 | |
| GSE77861 | SENCR | 100507392 | 239641_at | -0.1231 | 0.1454 | |
| SRP133303 | SENCR | 100507392 | RNAseq | -0.7030 | 0.0117 | |
| SRP159526 | SENCR | 100507392 | RNAseq | 0.8159 | 0.1266 |
Upregulated datasets: 0; Downregulated datasets: 0.
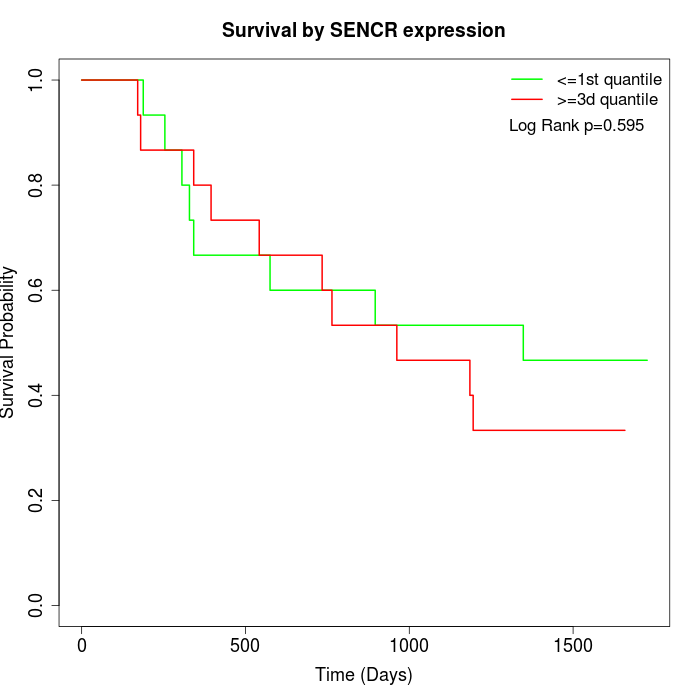
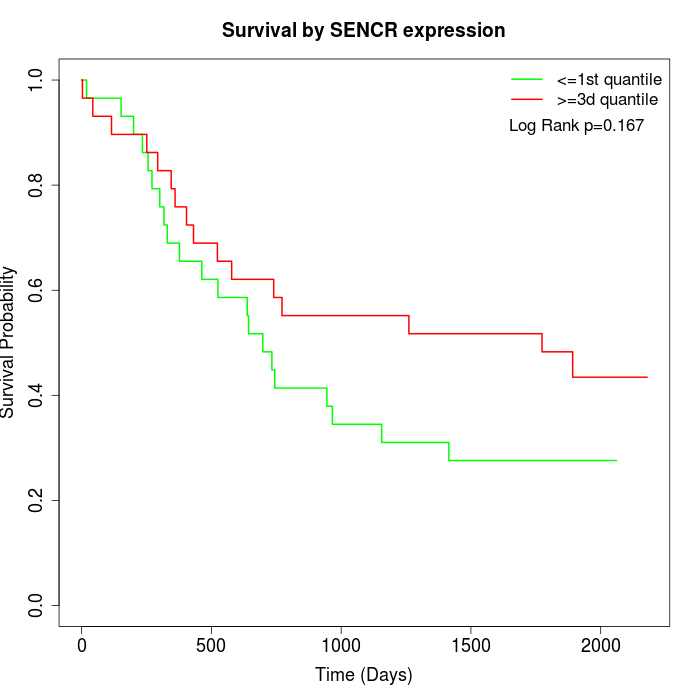
Survival by SENCR expression:
Note: Click image to view full size file.
Copy number change of SENCR:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SENCR | 100507392 | 0 | 14 | 16 | |
| GSE20123 | SENCR | 100507392 | 0 | 14 | 16 | |
| GSE43470 | SENCR | 100507392 | 2 | 7 | 34 | |
| GSE46452 | SENCR | 100507392 | 3 | 26 | 30 | |
| GSE47630 | SENCR | 100507392 | 2 | 19 | 19 | |
| GSE54993 | SENCR | 100507392 | 10 | 0 | 60 | |
| GSE54994 | SENCR | 100507392 | 5 | 19 | 29 | |
| GSE60625 | SENCR | 100507392 | 0 | 3 | 8 | |
| GSE74703 | SENCR | 100507392 | 1 | 5 | 30 | |
| GSE74704 | SENCR | 100507392 | 0 | 10 | 10 |
Total number of gains: 23; Total number of losses: 117; Total Number of normals: 252.
Somatic mutations of SENCR:
Generating mutation plots.
Highly correlated genes for SENCR:
Showing top 20/33 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SENCR | ZNF843 | 0.690621 | 3 | 0 | 3 |
| SENCR | C8B | 0.676867 | 3 | 0 | 3 |
| SENCR | APBB2 | 0.650569 | 3 | 0 | 3 |
| SENCR | EFCAB5 | 0.624064 | 3 | 0 | 3 |
| SENCR | TMEM31 | 0.623127 | 3 | 0 | 3 |
| SENCR | DNAJC5G | 0.616408 | 4 | 0 | 3 |
| SENCR | CDH22 | 0.613183 | 3 | 0 | 3 |
| SENCR | CAMKV | 0.606201 | 3 | 0 | 3 |
| SENCR | LINC00656 | 0.605375 | 4 | 0 | 3 |
| SENCR | EPOR | 0.601855 | 4 | 0 | 3 |
| SENCR | NR5A2 | 0.600532 | 3 | 0 | 3 |
| SENCR | CRYGA | 0.596557 | 3 | 0 | 3 |
| SENCR | PPP1R1A | 0.596339 | 4 | 0 | 3 |
| SENCR | TRIM10 | 0.594732 | 3 | 0 | 3 |
| SENCR | TMEM132B | 0.594108 | 3 | 0 | 3 |
| SENCR | CYGB | 0.590678 | 4 | 0 | 3 |
| SENCR | PCGEM1 | 0.585671 | 4 | 0 | 3 |
| SENCR | SIX3-AS1 | 0.581504 | 3 | 0 | 3 |
| SENCR | NMUR2 | 0.580834 | 4 | 0 | 3 |
| SENCR | PAX4 | 0.579781 | 4 | 0 | 3 |
For details and further investigation, click here