| Full name: crystallin gamma A | Alias Symbol: CRYG5|CRY-g-A | ||
| Type: protein-coding gene | Cytoband: 2q33.3 | ||
| Entrez ID: 1418 | HGNC ID: HGNC:2408 | Ensembl Gene: ENSG00000168582 | OMIM ID: 123660 |
| Drug and gene relationship at DGIdb | |||
Expression of CRYGA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CRYGA | 1418 | 207587_at | 0.0389 | 0.9062 | |
| GSE20347 | CRYGA | 1418 | 207587_at | -0.0148 | 0.8615 | |
| GSE23400 | CRYGA | 1418 | 207587_at | -0.0973 | 0.0162 | |
| GSE26886 | CRYGA | 1418 | 207587_at | 0.2110 | 0.0463 | |
| GSE29001 | CRYGA | 1418 | 207587_at | -0.2094 | 0.0595 | |
| GSE38129 | CRYGA | 1418 | 207587_at | -0.0808 | 0.2891 | |
| GSE45670 | CRYGA | 1418 | 207587_at | 0.0342 | 0.7744 | |
| GSE53622 | CRYGA | 1418 | 47599 | -0.3901 | 0.0000 | |
| GSE53624 | CRYGA | 1418 | 47599 | 0.1281 | 0.1753 | |
| GSE63941 | CRYGA | 1418 | 207587_at | 0.0552 | 0.7751 | |
| GSE77861 | CRYGA | 1418 | 207587_at | 0.0234 | 0.8689 | |
| GSE97050 | CRYGA | 1418 | A_23_P165380 | 0.1340 | 0.4678 |
Upregulated datasets: 0; Downregulated datasets: 0.
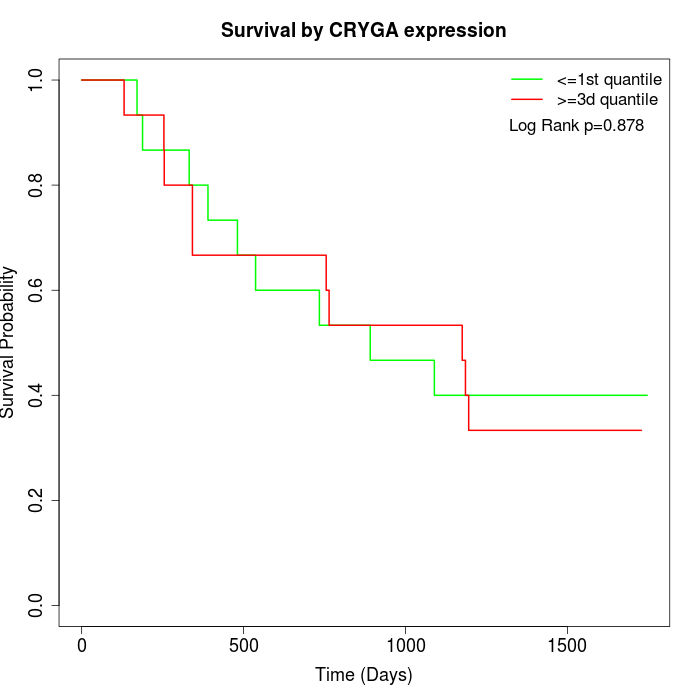
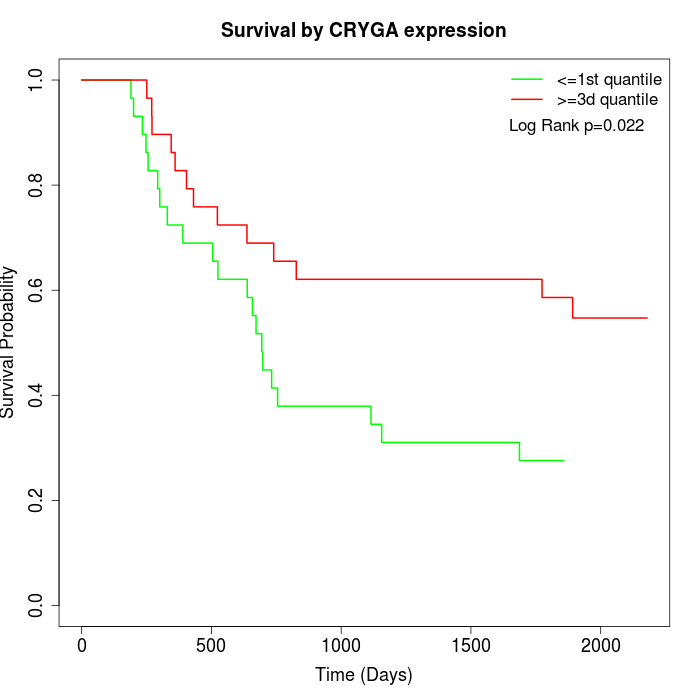
Survival by CRYGA expression:
Note: Click image to view full size file.
Copy number change of CRYGA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CRYGA | 1418 | 3 | 10 | 17 | |
| GSE20123 | CRYGA | 1418 | 3 | 10 | 17 | |
| GSE43470 | CRYGA | 1418 | 2 | 4 | 37 | |
| GSE46452 | CRYGA | 1418 | 0 | 5 | 54 | |
| GSE47630 | CRYGA | 1418 | 4 | 5 | 31 | |
| GSE54993 | CRYGA | 1418 | 0 | 3 | 67 | |
| GSE54994 | CRYGA | 1418 | 9 | 9 | 35 | |
| GSE60625 | CRYGA | 1418 | 0 | 3 | 8 | |
| GSE74703 | CRYGA | 1418 | 2 | 3 | 31 | |
| GSE74704 | CRYGA | 1418 | 2 | 5 | 13 | |
| TCGA | CRYGA | 1418 | 15 | 20 | 61 |
Total number of gains: 40; Total number of losses: 77; Total Number of normals: 371.
Somatic mutations of CRYGA:
Generating mutation plots.
Highly correlated genes for CRYGA:
Showing top 20/748 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CRYGA | TMEM179 | 0.77134 | 3 | 0 | 3 |
| CRYGA | KRTAP3-1 | 0.768776 | 3 | 0 | 3 |
| CRYGA | FAM131A | 0.767456 | 3 | 0 | 3 |
| CRYGA | SCN9A | 0.766809 | 3 | 0 | 3 |
| CRYGA | OR51T1 | 0.747451 | 3 | 0 | 3 |
| CRYGA | CALML6 | 0.744691 | 3 | 0 | 3 |
| CRYGA | KRTAP4-8 | 0.735987 | 3 | 0 | 3 |
| CRYGA | SALL3 | 0.731814 | 3 | 0 | 3 |
| CRYGA | SRD5A2 | 0.725362 | 4 | 0 | 3 |
| CRYGA | KRTAP6-1 | 0.724097 | 3 | 0 | 3 |
| CRYGA | SNX21 | 0.722058 | 3 | 0 | 3 |
| CRYGA | NTNG2 | 0.72136 | 3 | 0 | 3 |
| CRYGA | SP8 | 0.721045 | 4 | 0 | 4 |
| CRYGA | KRTAP4-1 | 0.718787 | 3 | 0 | 3 |
| CRYGA | SNED1 | 0.718244 | 4 | 0 | 4 |
| CRYGA | ARL4D | 0.716019 | 3 | 0 | 3 |
| CRYGA | PDE6C | 0.71551 | 4 | 0 | 4 |
| CRYGA | TPRX1 | 0.715227 | 3 | 0 | 3 |
| CRYGA | RASGRF2 | 0.714672 | 3 | 0 | 3 |
| CRYGA | ASCL1 | 0.714359 | 3 | 0 | 3 |
For details and further investigation, click here