| Full name: structural maintenance of chromosomes 1A | Alias Symbol: DXS423E|KIAA0178|SB1.8|Smcb | ||
| Type: protein-coding gene | Cytoband: Xp11.22 | ||
| Entrez ID: 8243 | HGNC ID: HGNC:11111 | Ensembl Gene: ENSG00000072501 | OMIM ID: 300040 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
SMC1A involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04110 | Cell cycle |
Expression of SMC1A:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SMC1A | 8243 | 201589_at | 0.6202 | 0.0867 | |
| GSE20347 | SMC1A | 8243 | 201589_at | 0.3387 | 0.0818 | |
| GSE23400 | SMC1A | 8243 | 201589_at | 0.6329 | 0.0000 | |
| GSE26886 | SMC1A | 8243 | 201589_at | 0.5838 | 0.0111 | |
| GSE29001 | SMC1A | 8243 | 201589_at | 0.5746 | 0.1556 | |
| GSE38129 | SMC1A | 8243 | 201589_at | 0.4101 | 0.0049 | |
| GSE45670 | SMC1A | 8243 | 201589_at | 0.2820 | 0.0893 | |
| GSE53622 | SMC1A | 8243 | 135412 | 0.1863 | 0.0028 | |
| GSE53624 | SMC1A | 8243 | 135412 | 0.6705 | 0.0000 | |
| GSE63941 | SMC1A | 8243 | 201589_at | 0.2609 | 0.4696 | |
| GSE77861 | SMC1A | 8243 | 201589_at | 0.3753 | 0.0925 | |
| GSE97050 | SMC1A | 8243 | A_33_P3290909 | 0.4979 | 0.1204 | |
| SRP007169 | SMC1A | 8243 | RNAseq | 2.4543 | 0.0000 | |
| SRP008496 | SMC1A | 8243 | RNAseq | 2.0973 | 0.0000 | |
| SRP064894 | SMC1A | 8243 | RNAseq | 0.4512 | 0.0761 | |
| SRP133303 | SMC1A | 8243 | RNAseq | 0.3232 | 0.1281 | |
| SRP159526 | SMC1A | 8243 | RNAseq | 0.5502 | 0.0187 | |
| SRP193095 | SMC1A | 8243 | RNAseq | 0.3308 | 0.0093 | |
| SRP219564 | SMC1A | 8243 | RNAseq | 0.4906 | 0.2141 | |
| TCGA | SMC1A | 8243 | RNAseq | 0.0441 | 0.3072 |
Upregulated datasets: 2; Downregulated datasets: 0.
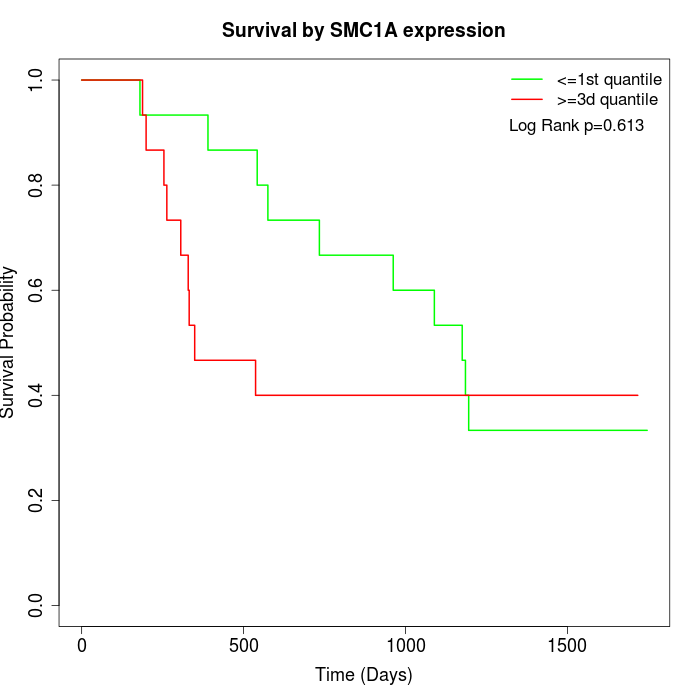
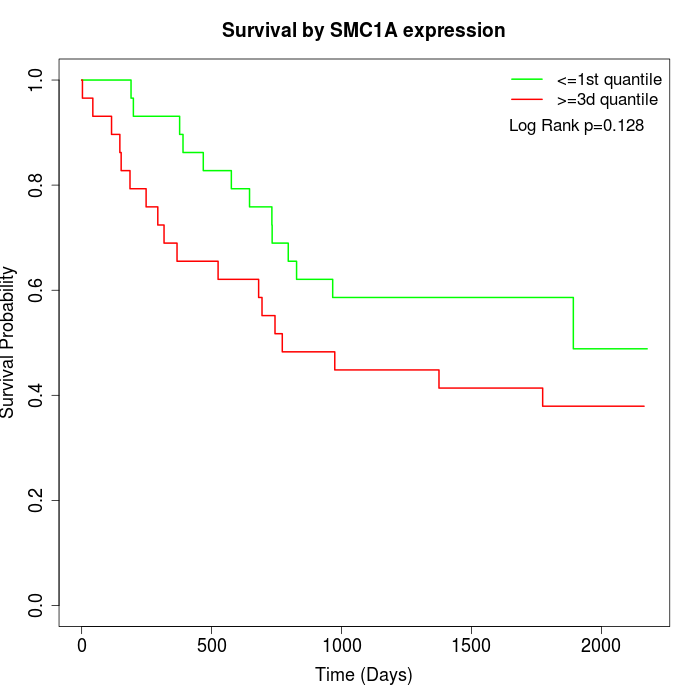
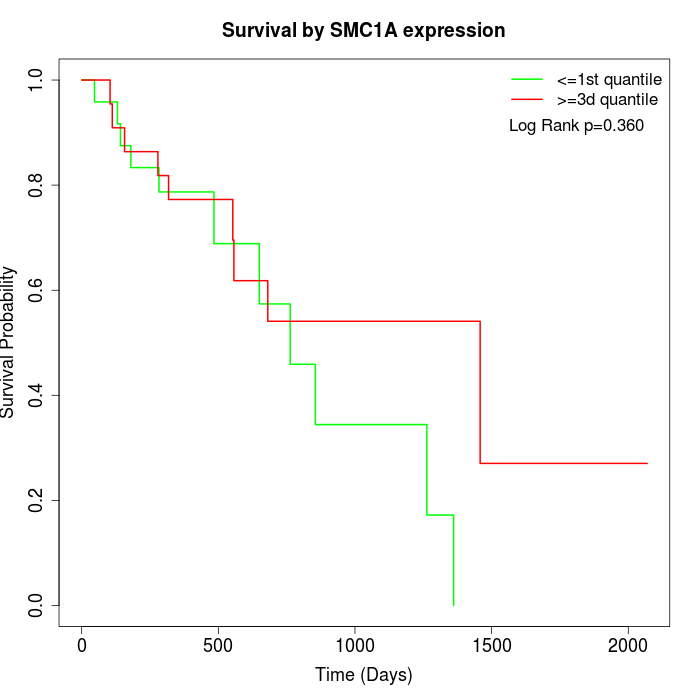
Survival by SMC1A expression:
Note: Click image to view full size file.
Copy number change of SMC1A:
No record found for this gene.
Somatic mutations of SMC1A:
Generating mutation plots.
Highly correlated genes for SMC1A:
Showing top 20/1128 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SMC1A | ZDHHC7 | 0.753788 | 3 | 0 | 3 |
| SMC1A | INTS10 | 0.730528 | 3 | 0 | 3 |
| SMC1A | ANKIB1 | 0.721571 | 3 | 0 | 3 |
| SMC1A | LRRC8B | 0.71801 | 3 | 0 | 3 |
| SMC1A | RCN2 | 0.690253 | 9 | 0 | 9 |
| SMC1A | SDHAF2 | 0.688076 | 3 | 0 | 3 |
| SMC1A | ASH2L | 0.686429 | 3 | 0 | 3 |
| SMC1A | FYN | 0.685789 | 3 | 0 | 3 |
| SMC1A | BIRC2 | 0.678426 | 4 | 0 | 4 |
| SMC1A | CYBA | 0.670879 | 3 | 0 | 3 |
| SMC1A | STX2 | 0.666229 | 3 | 0 | 3 |
| SMC1A | CDK4 | 0.664861 | 9 | 0 | 8 |
| SMC1A | NRM | 0.661193 | 4 | 0 | 3 |
| SMC1A | LARS2 | 0.659834 | 4 | 0 | 4 |
| SMC1A | TET1 | 0.65793 | 4 | 0 | 3 |
| SMC1A | NOL11 | 0.657872 | 9 | 0 | 8 |
| SMC1A | SLC35A2 | 0.656824 | 6 | 0 | 6 |
| SMC1A | SMCR8 | 0.655993 | 4 | 0 | 3 |
| SMC1A | UBTD2 | 0.652686 | 4 | 0 | 4 |
| SMC1A | ELAVL1 | 0.652431 | 8 | 0 | 8 |
For details and further investigation, click here