| Full name: ELAV like RNA binding protein 1 | Alias Symbol: HuR|Hua|MelG | ||
| Type: protein-coding gene | Cytoband: 19p13.2 | ||
| Entrez ID: 1994 | HGNC ID: HGNC:3312 | Ensembl Gene: ENSG00000066044 | OMIM ID: 603466 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ELAVL1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04152 | AMPK signaling pathway |
Expression of ELAVL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ELAVL1 | 1994 | 201726_at | 0.5428 | 0.1007 | |
| GSE20347 | ELAVL1 | 1994 | 201726_at | 0.4230 | 0.0139 | |
| GSE23400 | ELAVL1 | 1994 | 201726_at | 0.5975 | 0.0000 | |
| GSE26886 | ELAVL1 | 1994 | 201726_at | -0.6238 | 0.0001 | |
| GSE29001 | ELAVL1 | 1994 | 201726_at | 0.4959 | 0.0812 | |
| GSE38129 | ELAVL1 | 1994 | 201726_at | 0.4822 | 0.0004 | |
| GSE45670 | ELAVL1 | 1994 | 201726_at | 0.2455 | 0.0115 | |
| GSE53622 | ELAVL1 | 1994 | 17038 | 0.1501 | 0.0081 | |
| GSE53624 | ELAVL1 | 1994 | 17038 | 0.4552 | 0.0000 | |
| GSE63941 | ELAVL1 | 1994 | 201726_at | 0.1667 | 0.5105 | |
| GSE77861 | ELAVL1 | 1994 | 201726_at | 0.4050 | 0.0257 | |
| GSE97050 | ELAVL1 | 1994 | A_23_P388681 | 0.1205 | 0.7234 | |
| SRP007169 | ELAVL1 | 1994 | RNAseq | 0.9361 | 0.0056 | |
| SRP008496 | ELAVL1 | 1994 | RNAseq | 1.0235 | 0.0000 | |
| SRP064894 | ELAVL1 | 1994 | RNAseq | 0.4093 | 0.0011 | |
| SRP133303 | ELAVL1 | 1994 | RNAseq | 0.3135 | 0.0246 | |
| SRP159526 | ELAVL1 | 1994 | RNAseq | 0.5722 | 0.0016 | |
| SRP193095 | ELAVL1 | 1994 | RNAseq | 0.3517 | 0.0000 | |
| SRP219564 | ELAVL1 | 1994 | RNAseq | 0.5316 | 0.0493 | |
| TCGA | ELAVL1 | 1994 | RNAseq | 0.1285 | 0.0046 |
Upregulated datasets: 1; Downregulated datasets: 0.
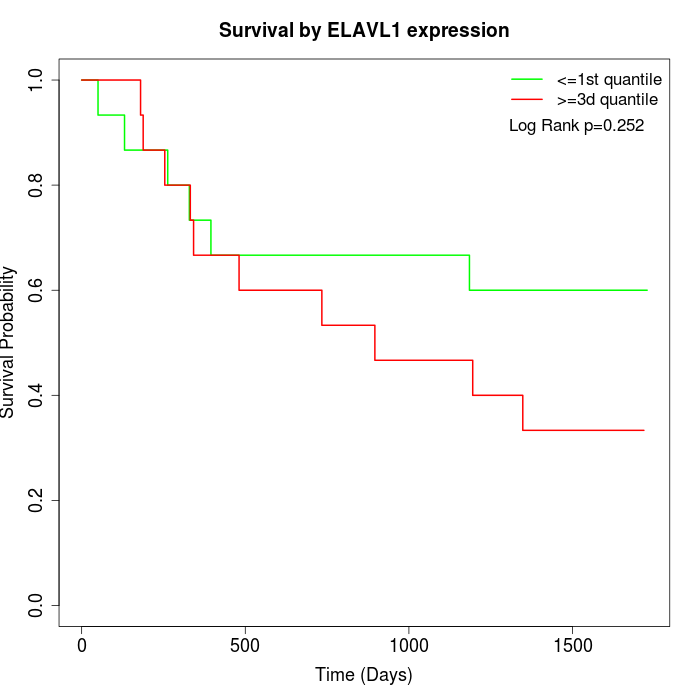
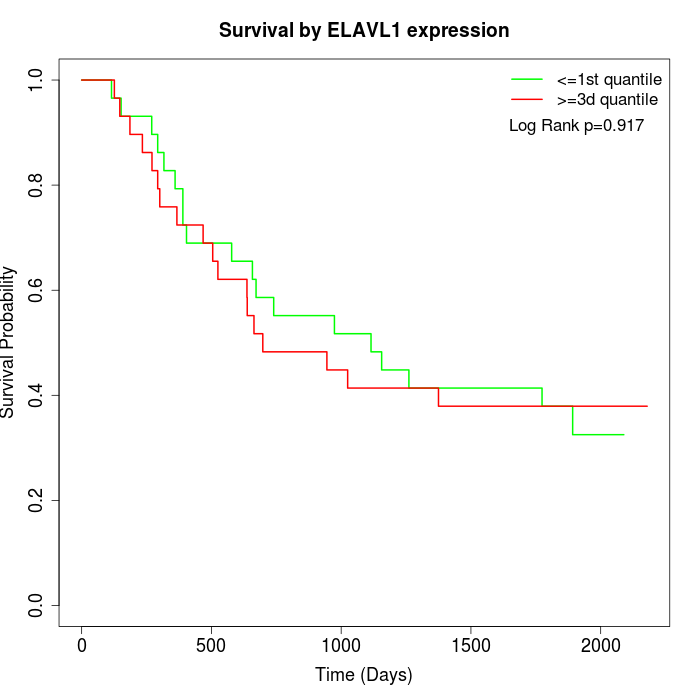
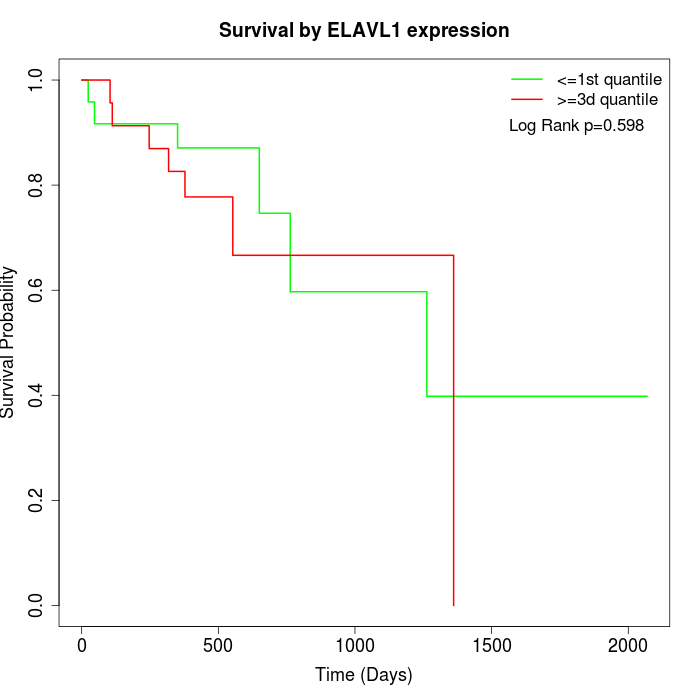
Survival by ELAVL1 expression:
Note: Click image to view full size file.
Copy number change of ELAVL1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ELAVL1 | 1994 | 4 | 3 | 23 | |
| GSE20123 | ELAVL1 | 1994 | 3 | 2 | 25 | |
| GSE43470 | ELAVL1 | 1994 | 2 | 7 | 34 | |
| GSE46452 | ELAVL1 | 1994 | 47 | 1 | 11 | |
| GSE47630 | ELAVL1 | 1994 | 5 | 7 | 28 | |
| GSE54993 | ELAVL1 | 1994 | 16 | 3 | 51 | |
| GSE54994 | ELAVL1 | 1994 | 6 | 14 | 33 | |
| GSE60625 | ELAVL1 | 1994 | 9 | 0 | 2 | |
| GSE74703 | ELAVL1 | 1994 | 2 | 4 | 30 | |
| GSE74704 | ELAVL1 | 1994 | 0 | 2 | 18 | |
| TCGA | ELAVL1 | 1994 | 10 | 20 | 66 |
Total number of gains: 104; Total number of losses: 63; Total Number of normals: 321.
Somatic mutations of ELAVL1:
Generating mutation plots.
Highly correlated genes for ELAVL1:
Showing top 20/1134 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ELAVL1 | MRPL38 | 0.803069 | 3 | 0 | 3 |
| ELAVL1 | SYK | 0.792004 | 3 | 0 | 3 |
| ELAVL1 | HPS3 | 0.786973 | 4 | 0 | 4 |
| ELAVL1 | ARF3 | 0.776448 | 3 | 0 | 3 |
| ELAVL1 | AKT1S1 | 0.774951 | 3 | 0 | 3 |
| ELAVL1 | CWF19L1 | 0.773008 | 3 | 0 | 3 |
| ELAVL1 | ZNF317 | 0.760221 | 5 | 0 | 5 |
| ELAVL1 | PRMT6 | 0.755115 | 3 | 0 | 3 |
| ELAVL1 | DNAJC9 | 0.750587 | 7 | 0 | 7 |
| ELAVL1 | ORAI1 | 0.748742 | 4 | 0 | 4 |
| ELAVL1 | RNPS1 | 0.748073 | 7 | 0 | 7 |
| ELAVL1 | CERS5 | 0.741755 | 3 | 0 | 3 |
| ELAVL1 | XPO1 | 0.734339 | 9 | 0 | 9 |
| ELAVL1 | ISY1 | 0.7262 | 3 | 0 | 3 |
| ELAVL1 | MRPL3 | 0.723814 | 7 | 0 | 7 |
| ELAVL1 | RSRC2 | 0.723074 | 3 | 0 | 3 |
| ELAVL1 | MSH6 | 0.720042 | 10 | 0 | 8 |
| ELAVL1 | HMGN2 | 0.718765 | 5 | 0 | 5 |
| ELAVL1 | DDX50 | 0.712506 | 6 | 0 | 6 |
| ELAVL1 | TUBA1B | 0.712494 | 9 | 0 | 7 |
For details and further investigation, click here