| Full name: sphingomyelin phosphodiesterase 4 | Alias Symbol: FLJ20297|FLJ20756|nSMase-3|KIAA1418|NSMASE3|NET13 | ||
| Type: protein-coding gene | Cytoband: 2q21.1 | ||
| Entrez ID: 55627 | HGNC ID: HGNC:32949 | Ensembl Gene: ENSG00000136699 | OMIM ID: 610457 |
| Drug and gene relationship at DGIdb | |||
Expression of SMPD4:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | SMPD4 | 55627 | 71430 | 0.5786 | 0.0000 | |
| GSE53624 | SMPD4 | 55627 | 71430 | 0.6420 | 0.0000 | |
| GSE97050 | SMPD4 | 55627 | A_23_P255286 | 0.1780 | 0.3987 | |
| SRP007169 | SMPD4 | 55627 | RNAseq | 0.8998 | 0.0049 | |
| SRP008496 | SMPD4 | 55627 | RNAseq | 0.7867 | 0.0000 | |
| SRP064894 | SMPD4 | 55627 | RNAseq | 0.6498 | 0.0000 | |
| SRP133303 | SMPD4 | 55627 | RNAseq | 0.6772 | 0.0001 | |
| SRP159526 | SMPD4 | 55627 | RNAseq | 0.8285 | 0.0130 | |
| SRP193095 | SMPD4 | 55627 | RNAseq | 0.6610 | 0.0000 | |
| SRP219564 | SMPD4 | 55627 | RNAseq | 0.3130 | 0.2316 | |
| TCGA | SMPD4 | 55627 | RNAseq | 0.1535 | 0.0005 |
Upregulated datasets: 0; Downregulated datasets: 0.
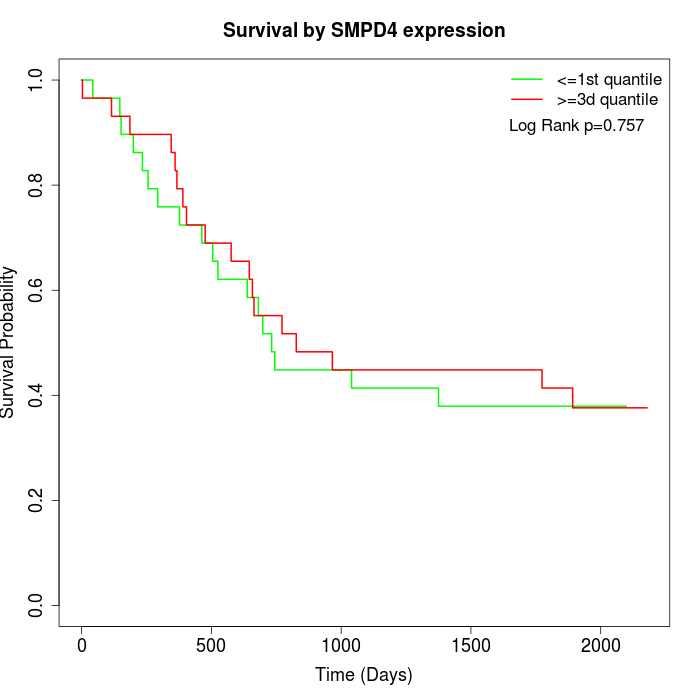
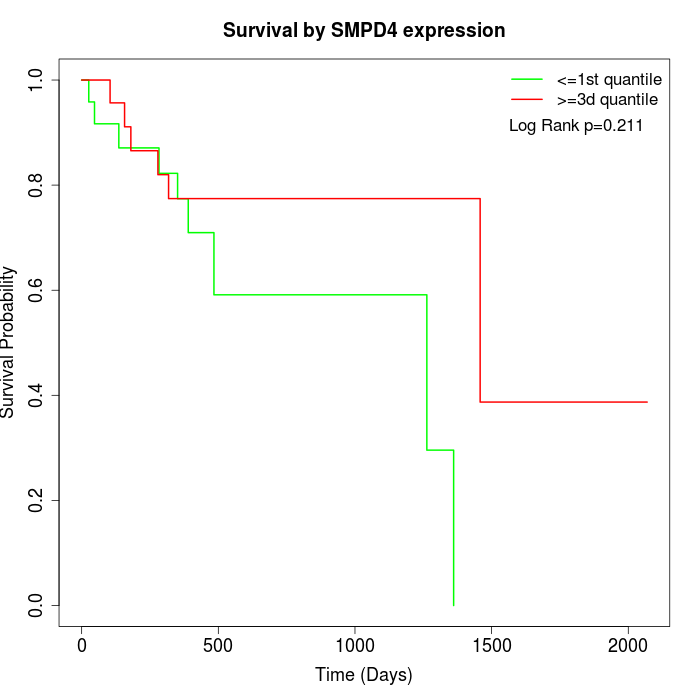
Survival by SMPD4 expression:
Note: Click image to view full size file.
Copy number change of SMPD4:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SMPD4 | 55627 | 5 | 3 | 22 | |
| GSE20123 | SMPD4 | 55627 | 5 | 3 | 22 | |
| GSE43470 | SMPD4 | 55627 | 2 | 2 | 39 | |
| GSE46452 | SMPD4 | 55627 | 1 | 3 | 55 | |
| GSE47630 | SMPD4 | 55627 | 5 | 0 | 35 | |
| GSE54993 | SMPD4 | 55627 | 0 | 6 | 64 | |
| GSE54994 | SMPD4 | 55627 | 11 | 0 | 42 | |
| GSE60625 | SMPD4 | 55627 | 0 | 3 | 8 | |
| GSE74703 | SMPD4 | 55627 | 2 | 1 | 33 | |
| GSE74704 | SMPD4 | 55627 | 3 | 1 | 16 | |
| TCGA | SMPD4 | 55627 | 22 | 9 | 65 |
Total number of gains: 56; Total number of losses: 31; Total Number of normals: 401.
Somatic mutations of SMPD4:
Generating mutation plots.
Highly correlated genes for SMPD4:
Showing top 20/55 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SMPD4 | CDCA5 | 0.80049 | 3 | 0 | 3 |
| SMPD4 | KIF19 | 0.78074 | 3 | 0 | 3 |
| SMPD4 | YEATS2 | 0.772992 | 3 | 0 | 3 |
| SMPD4 | LRFN4 | 0.757555 | 3 | 0 | 3 |
| SMPD4 | E2F1 | 0.749352 | 3 | 0 | 3 |
| SMPD4 | KIF4A | 0.738645 | 3 | 0 | 3 |
| SMPD4 | GINS2 | 0.733564 | 3 | 0 | 3 |
| SMPD4 | DBF4B | 0.721882 | 3 | 0 | 3 |
| SMPD4 | CBX3 | 0.72048 | 3 | 0 | 3 |
| SMPD4 | FAM189B | 0.718419 | 3 | 0 | 3 |
| SMPD4 | CBX8 | 0.714037 | 3 | 0 | 3 |
| SMPD4 | DNMT3B | 0.709896 | 3 | 0 | 3 |
| SMPD4 | SMG5 | 0.705027 | 3 | 0 | 3 |
| SMPD4 | ATAD3C | 0.701408 | 3 | 0 | 3 |
| SMPD4 | C1QTNF6 | 0.696572 | 3 | 0 | 3 |
| SMPD4 | GPRIN1 | 0.695774 | 3 | 0 | 3 |
| SMPD4 | COL27A1 | 0.691327 | 3 | 0 | 3 |
| SMPD4 | KAT2A | 0.690727 | 3 | 0 | 3 |
| SMPD4 | MDC1 | 0.689692 | 3 | 0 | 3 |
| SMPD4 | ODF2 | 0.688028 | 3 | 0 | 3 |
For details and further investigation, click here