| Full name: SMAD specific E3 ubiquitin protein ligase 2 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 17q23.3-q24.1 | ||
| Entrez ID: 64750 | HGNC ID: HGNC:16809 | Ensembl Gene: ENSG00000108854 | OMIM ID: 605532 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
SMURF2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04340 | Hedgehog signaling pathway | |
| hsa04350 | TGF-beta signaling pathway |
Expression of SMURF2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SMURF2 | 64750 | 205596_s_at | 0.2170 | 0.6051 | |
| GSE20347 | SMURF2 | 64750 | 205596_s_at | 0.4870 | 0.0049 | |
| GSE23400 | SMURF2 | 64750 | 205596_s_at | 0.7217 | 0.0000 | |
| GSE26886 | SMURF2 | 64750 | 205596_s_at | 1.7854 | 0.0000 | |
| GSE29001 | SMURF2 | 64750 | 205596_s_at | 0.6985 | 0.0303 | |
| GSE38129 | SMURF2 | 64750 | 205596_s_at | 0.4244 | 0.0017 | |
| GSE45670 | SMURF2 | 64750 | 205596_s_at | 0.1830 | 0.5007 | |
| GSE53622 | SMURF2 | 64750 | 155590 | 0.0831 | 0.4999 | |
| GSE53624 | SMURF2 | 64750 | 155590 | 0.1475 | 0.3835 | |
| GSE63941 | SMURF2 | 64750 | 205596_s_at | -1.9417 | 0.0123 | |
| GSE77861 | SMURF2 | 64750 | 205596_s_at | 1.3720 | 0.0079 | |
| GSE97050 | SMURF2 | 64750 | A_33_P3370751 | -0.0775 | 0.7333 | |
| SRP007169 | SMURF2 | 64750 | RNAseq | 0.8748 | 0.0647 | |
| SRP008496 | SMURF2 | 64750 | RNAseq | 1.0027 | 0.0003 | |
| SRP064894 | SMURF2 | 64750 | RNAseq | 0.2499 | 0.1842 | |
| SRP133303 | SMURF2 | 64750 | RNAseq | 0.8349 | 0.0000 | |
| SRP159526 | SMURF2 | 64750 | RNAseq | 0.7360 | 0.0012 | |
| SRP193095 | SMURF2 | 64750 | RNAseq | 0.4727 | 0.0002 | |
| SRP219564 | SMURF2 | 64750 | RNAseq | 0.3492 | 0.1532 | |
| TCGA | SMURF2 | 64750 | RNAseq | -0.0177 | 0.7493 |
Upregulated datasets: 3; Downregulated datasets: 1.
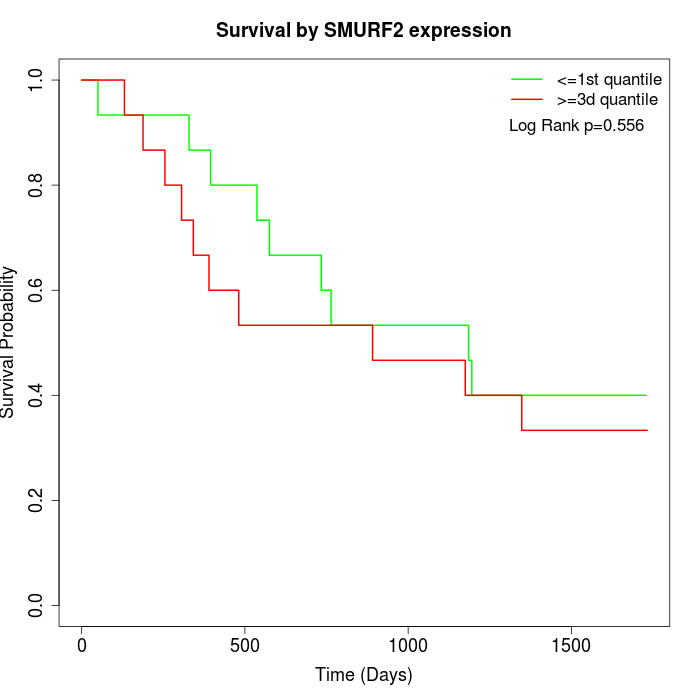
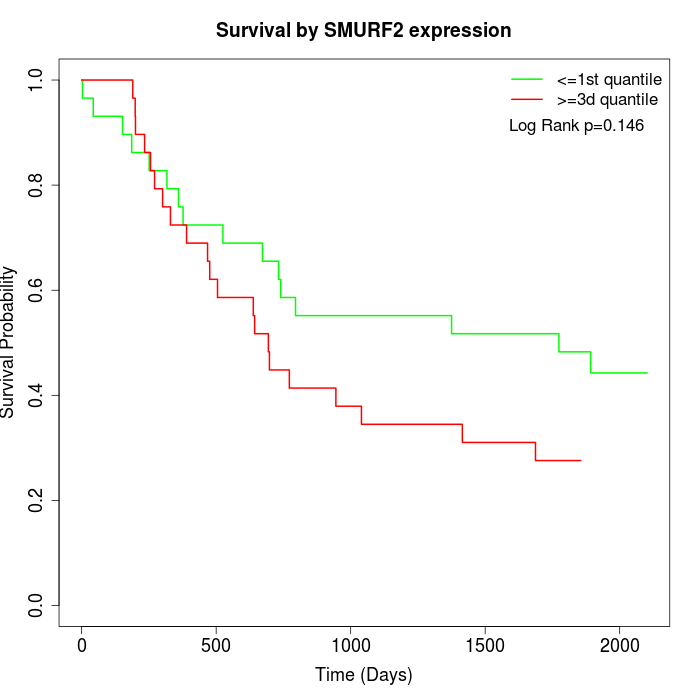
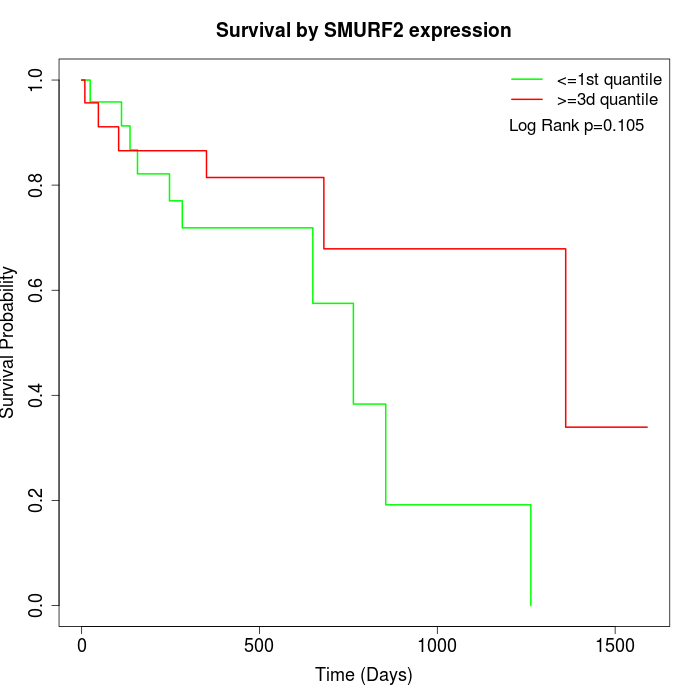
Survival by SMURF2 expression:
Note: Click image to view full size file.
Copy number change of SMURF2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SMURF2 | 64750 | 3 | 1 | 26 | |
| GSE20123 | SMURF2 | 64750 | 3 | 1 | 26 | |
| GSE43470 | SMURF2 | 64750 | 6 | 0 | 37 | |
| GSE46452 | SMURF2 | 64750 | 32 | 0 | 27 | |
| GSE47630 | SMURF2 | 64750 | 7 | 1 | 32 | |
| GSE54993 | SMURF2 | 64750 | 2 | 5 | 63 | |
| GSE54994 | SMURF2 | 64750 | 9 | 4 | 40 | |
| GSE60625 | SMURF2 | 64750 | 4 | 0 | 7 | |
| GSE74703 | SMURF2 | 64750 | 6 | 0 | 30 | |
| GSE74704 | SMURF2 | 64750 | 3 | 1 | 16 | |
| TCGA | SMURF2 | 64750 | 30 | 8 | 58 |
Total number of gains: 105; Total number of losses: 21; Total Number of normals: 362.
Somatic mutations of SMURF2:
Generating mutation plots.
Highly correlated genes for SMURF2:
Showing top 20/1513 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SMURF2 | IGFBP5 | 0.795788 | 3 | 0 | 3 |
| SMURF2 | CCNT1 | 0.765852 | 3 | 0 | 3 |
| SMURF2 | METTL8 | 0.746169 | 3 | 0 | 3 |
| SMURF2 | RDH14 | 0.744793 | 3 | 0 | 3 |
| SMURF2 | KCTD7 | 0.737088 | 3 | 0 | 3 |
| SMURF2 | TTC7B | 0.736919 | 6 | 0 | 6 |
| SMURF2 | LINC00674 | 0.73056 | 4 | 0 | 3 |
| SMURF2 | PHIP | 0.725946 | 5 | 0 | 5 |
| SMURF2 | PGAP1 | 0.725072 | 3 | 0 | 3 |
| SMURF2 | RNF217 | 0.724532 | 3 | 0 | 3 |
| SMURF2 | BIVM | 0.723675 | 3 | 0 | 3 |
| SMURF2 | RILPL1 | 0.720527 | 4 | 0 | 4 |
| SMURF2 | RNASEH2C | 0.720241 | 4 | 0 | 3 |
| SMURF2 | HABP4 | 0.717711 | 4 | 0 | 3 |
| SMURF2 | PCDH18 | 0.706915 | 5 | 0 | 4 |
| SMURF2 | IFT22 | 0.706158 | 3 | 0 | 3 |
| SMURF2 | IWS1 | 0.703747 | 4 | 0 | 3 |
| SMURF2 | ZNF644 | 0.70363 | 3 | 0 | 3 |
| SMURF2 | GLS | 0.700842 | 7 | 0 | 6 |
| SMURF2 | RRP15 | 0.697848 | 8 | 0 | 8 |
For details and further investigation, click here