| Full name: retinol dehydrogenase 14 | Alias Symbol: PAN2|SDR7C4 | ||
| Type: protein-coding gene | Cytoband: 2p24.2 | ||
| Entrez ID: 57665 | HGNC ID: HGNC:19979 | Ensembl Gene: ENSG00000240857 | OMIM ID: 616796 |
| Drug and gene relationship at DGIdb | |||
Expression of RDH14:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | RDH14 | 57665 | 90853 | 0.0558 | 0.4673 | |
| GSE53624 | RDH14 | 57665 | 90853 | 0.1584 | 0.0180 | |
| GSE97050 | RDH14 | 57665 | A_23_P90911 | -0.1540 | 0.6814 | |
| SRP007169 | RDH14 | 57665 | RNAseq | -0.0455 | 0.9111 | |
| SRP008496 | RDH14 | 57665 | RNAseq | 0.4713 | 0.1011 | |
| SRP064894 | RDH14 | 57665 | RNAseq | 0.2067 | 0.2133 | |
| SRP133303 | RDH14 | 57665 | RNAseq | 0.3533 | 0.0077 | |
| SRP159526 | RDH14 | 57665 | RNAseq | 0.5994 | 0.0441 | |
| SRP193095 | RDH14 | 57665 | RNAseq | 0.0073 | 0.9601 | |
| SRP219564 | RDH14 | 57665 | RNAseq | 0.2461 | 0.5120 | |
| TCGA | RDH14 | 57665 | RNAseq | -0.0577 | 0.3182 |
Upregulated datasets: 0; Downregulated datasets: 0.
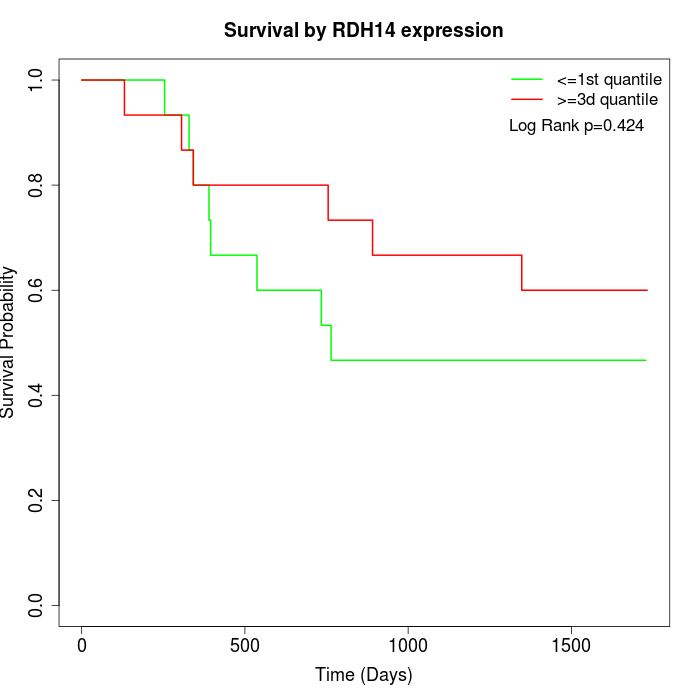
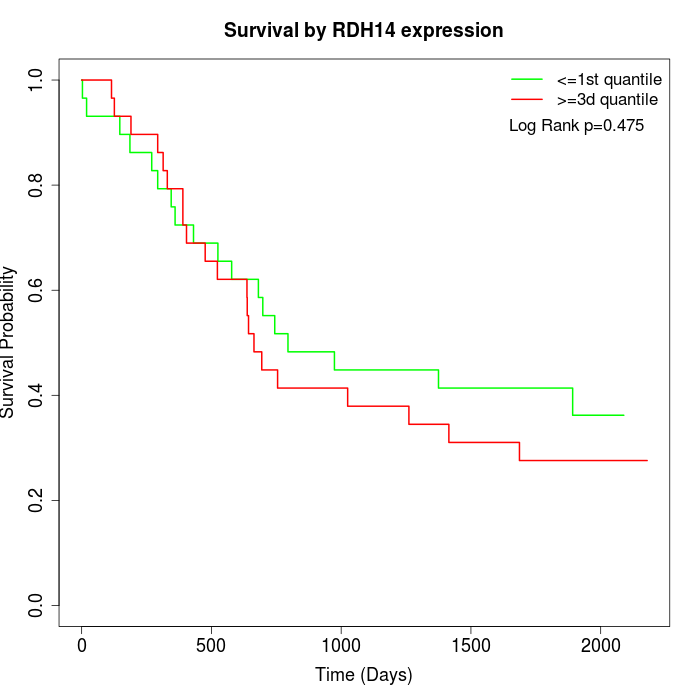
Survival by RDH14 expression:
Note: Click image to view full size file.
Copy number change of RDH14:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | RDH14 | 57665 | 10 | 2 | 18 | |
| GSE20123 | RDH14 | 57665 | 10 | 2 | 18 | |
| GSE43470 | RDH14 | 57665 | 2 | 0 | 41 | |
| GSE46452 | RDH14 | 57665 | 3 | 4 | 52 | |
| GSE47630 | RDH14 | 57665 | 7 | 0 | 33 | |
| GSE54993 | RDH14 | 57665 | 1 | 7 | 62 | |
| GSE54994 | RDH14 | 57665 | 10 | 1 | 42 | |
| GSE60625 | RDH14 | 57665 | 0 | 3 | 8 | |
| GSE74703 | RDH14 | 57665 | 2 | 0 | 34 | |
| GSE74704 | RDH14 | 57665 | 9 | 2 | 9 | |
| TCGA | RDH14 | 57665 | 36 | 3 | 57 |
Total number of gains: 90; Total number of losses: 24; Total Number of normals: 374.
Somatic mutations of RDH14:
Generating mutation plots.
Highly correlated genes for RDH14:
Showing top 20/26 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| RDH14 | MRPL33 | 0.787983 | 3 | 0 | 3 |
| RDH14 | MAP4K3 | 0.74879 | 3 | 0 | 3 |
| RDH14 | SMURF2 | 0.744793 | 3 | 0 | 3 |
| RDH14 | ZNF512 | 0.736285 | 3 | 0 | 3 |
| RDH14 | HELZ | 0.714333 | 3 | 0 | 3 |
| RDH14 | UBR1 | 0.712551 | 3 | 0 | 3 |
| RDH14 | VPS13C | 0.711822 | 3 | 0 | 3 |
| RDH14 | DYNC2LI1 | 0.709896 | 3 | 0 | 3 |
| RDH14 | THAP1 | 0.708668 | 3 | 0 | 3 |
| RDH14 | IFT81 | 0.708542 | 3 | 0 | 3 |
| RDH14 | PLEKHA3 | 0.703916 | 3 | 0 | 3 |
| RDH14 | ABI2 | 0.700192 | 3 | 0 | 3 |
| RDH14 | PCGF6 | 0.694303 | 3 | 0 | 3 |
| RDH14 | PGRMC1 | 0.692667 | 3 | 0 | 3 |
| RDH14 | CD164 | 0.675648 | 3 | 0 | 3 |
| RDH14 | LYSMD1 | 0.672847 | 3 | 0 | 3 |
| RDH14 | DMXL2 | 0.668646 | 3 | 0 | 3 |
| RDH14 | HEATR6 | 0.659182 | 3 | 0 | 3 |
| RDH14 | PREPL | 0.653243 | 3 | 0 | 3 |
| RDH14 | BMI1 | 0.650788 | 4 | 0 | 3 |
For details and further investigation, click here