| Full name: post-GPI attachment to proteins 1 | Alias Symbol: FLJ12377|Bst1|SPG67 | ||
| Type: protein-coding gene | Cytoband: 2q33.1 | ||
| Entrez ID: 80055 | HGNC ID: HGNC:25712 | Ensembl Gene: ENSG00000197121 | OMIM ID: 611655 |
| Drug and gene relationship at DGIdb | |||
Expression of PGAP1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PGAP1 | 80055 | 220576_at | 0.0842 | 0.8847 | |
| GSE20347 | PGAP1 | 80055 | 220576_at | 0.5389 | 0.0497 | |
| GSE23400 | PGAP1 | 80055 | 220576_at | 0.4250 | 0.0000 | |
| GSE26886 | PGAP1 | 80055 | 213469_at | 1.5001 | 0.0007 | |
| GSE29001 | PGAP1 | 80055 | 220576_at | 0.0697 | 0.8858 | |
| GSE38129 | PGAP1 | 80055 | 220576_at | 0.6920 | 0.0375 | |
| GSE45670 | PGAP1 | 80055 | 213469_at | 0.4168 | 0.1859 | |
| GSE53622 | PGAP1 | 80055 | 53997 | 0.5878 | 0.0000 | |
| GSE53624 | PGAP1 | 80055 | 53997 | 0.5972 | 0.0000 | |
| GSE63941 | PGAP1 | 80055 | 213469_at | -0.1445 | 0.8378 | |
| GSE77861 | PGAP1 | 80055 | 220576_at | 0.8011 | 0.0269 | |
| GSE97050 | PGAP1 | 80055 | A_23_P79247 | 0.1429 | 0.6456 | |
| SRP007169 | PGAP1 | 80055 | RNAseq | 1.4877 | 0.0084 | |
| SRP008496 | PGAP1 | 80055 | RNAseq | 1.2435 | 0.0003 | |
| SRP064894 | PGAP1 | 80055 | RNAseq | -0.0622 | 0.8439 | |
| SRP133303 | PGAP1 | 80055 | RNAseq | 0.3372 | 0.0995 | |
| SRP159526 | PGAP1 | 80055 | RNAseq | 0.2048 | 0.3580 | |
| SRP193095 | PGAP1 | 80055 | RNAseq | -0.0583 | 0.6477 | |
| SRP219564 | PGAP1 | 80055 | RNAseq | -0.1295 | 0.8037 | |
| TCGA | PGAP1 | 80055 | RNAseq | 0.1580 | 0.0595 |
Upregulated datasets: 3; Downregulated datasets: 0.
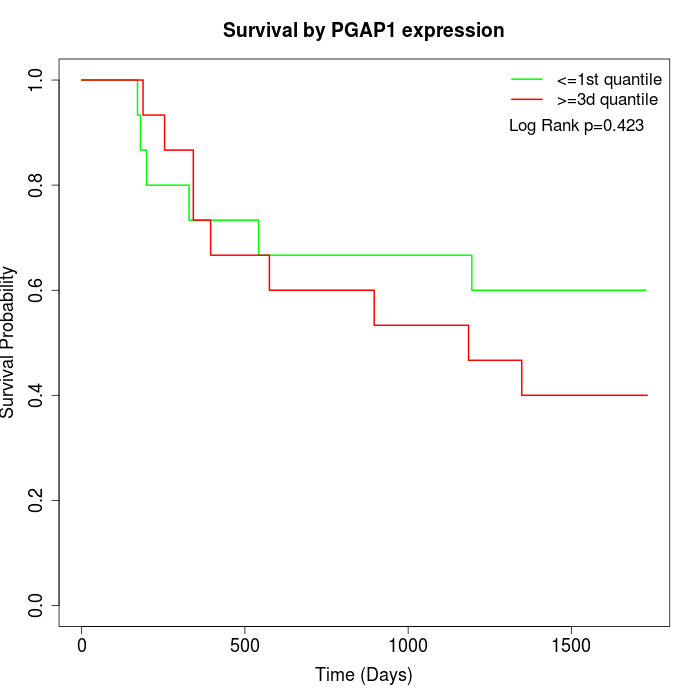
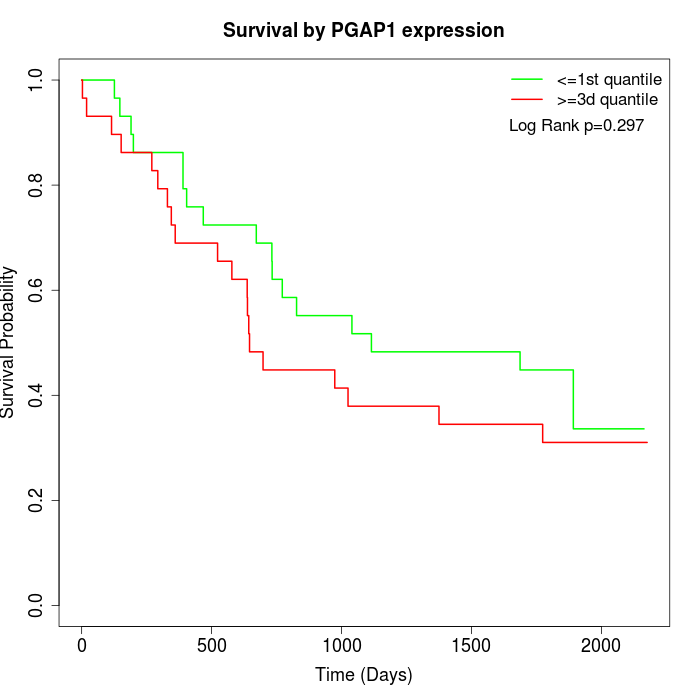
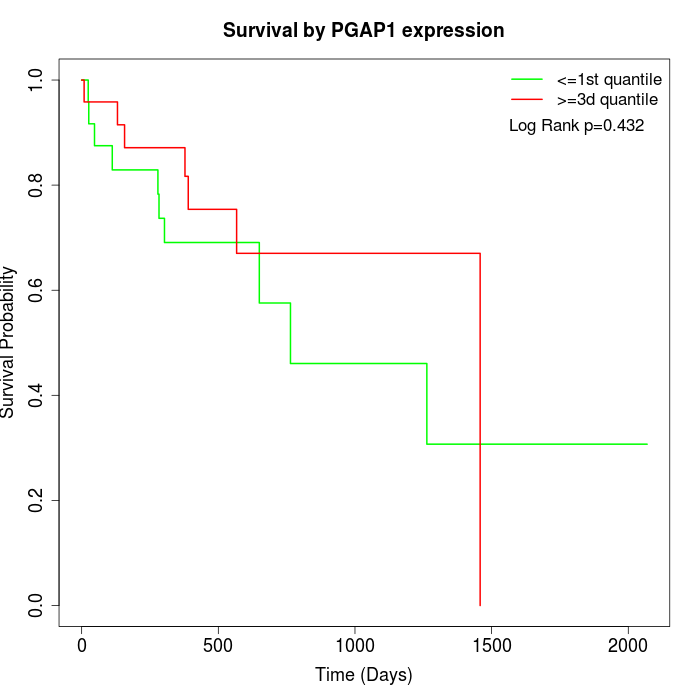
Survival by PGAP1 expression:
Note: Click image to view full size file.
Copy number change of PGAP1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PGAP1 | 80055 | 10 | 2 | 18 | |
| GSE20123 | PGAP1 | 80055 | 10 | 2 | 18 | |
| GSE43470 | PGAP1 | 80055 | 3 | 1 | 39 | |
| GSE46452 | PGAP1 | 80055 | 1 | 4 | 54 | |
| GSE47630 | PGAP1 | 80055 | 4 | 5 | 31 | |
| GSE54993 | PGAP1 | 80055 | 0 | 4 | 66 | |
| GSE54994 | PGAP1 | 80055 | 11 | 7 | 35 | |
| GSE60625 | PGAP1 | 80055 | 0 | 3 | 8 | |
| GSE74703 | PGAP1 | 80055 | 2 | 1 | 33 | |
| GSE74704 | PGAP1 | 80055 | 5 | 2 | 13 | |
| TCGA | PGAP1 | 80055 | 22 | 9 | 65 |
Total number of gains: 68; Total number of losses: 40; Total Number of normals: 380.
Somatic mutations of PGAP1:
Generating mutation plots.
Highly correlated genes for PGAP1:
Showing top 20/712 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PGAP1 | RPF2 | 0.839971 | 3 | 0 | 3 |
| PGAP1 | ZCCHC7 | 0.797672 | 3 | 0 | 3 |
| PGAP1 | NAP1L1 | 0.779754 | 3 | 0 | 3 |
| PGAP1 | ZBED6 | 0.77339 | 3 | 0 | 3 |
| PGAP1 | EPN1 | 0.767238 | 3 | 0 | 3 |
| PGAP1 | MAK16 | 0.766669 | 3 | 0 | 3 |
| PGAP1 | GOPC | 0.763632 | 3 | 0 | 3 |
| PGAP1 | NKTR | 0.762333 | 3 | 0 | 3 |
| PGAP1 | MRPL52 | 0.756885 | 3 | 0 | 3 |
| PGAP1 | DDX59 | 0.74963 | 4 | 0 | 3 |
| PGAP1 | FAM161B | 0.738599 | 3 | 0 | 3 |
| PGAP1 | C9orf64 | 0.731449 | 4 | 0 | 3 |
| PGAP1 | GLS | 0.730329 | 3 | 0 | 3 |
| PGAP1 | ZNF251 | 0.727995 | 4 | 0 | 4 |
| PGAP1 | SPDYE3 | 0.727139 | 3 | 0 | 3 |
| PGAP1 | TMED8 | 0.726184 | 3 | 0 | 3 |
| PGAP1 | SMURF2 | 0.725072 | 3 | 0 | 3 |
| PGAP1 | LYAR | 0.722973 | 3 | 0 | 3 |
| PGAP1 | TKT | 0.711708 | 3 | 0 | 3 |
| PGAP1 | CPSF3 | 0.709356 | 3 | 0 | 3 |
For details and further investigation, click here