| Full name: potassium channel tetramerization domain containing 7 | Alias Symbol: FLJ32069|EPM3|CLN14 | ||
| Type: protein-coding gene | Cytoband: 7q11.21 | ||
| Entrez ID: 154881 | HGNC ID: HGNC:21957 | Ensembl Gene: ENSG00000243335 | OMIM ID: 611725 |
| Drug and gene relationship at DGIdb | |||
Expression of KCTD7:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | KCTD7 | 154881 | 26729 | 0.0686 | 0.6738 | |
| GSE53624 | KCTD7 | 154881 | 26729 | 0.2799 | 0.0443 | |
| GSE97050 | KCTD7 | 154881 | A_24_P329229 | -0.1915 | 0.5501 | |
| SRP007169 | KCTD7 | 154881 | RNAseq | 0.6891 | 0.0986 | |
| SRP008496 | KCTD7 | 154881 | RNAseq | 0.6459 | 0.0341 | |
| SRP064894 | KCTD7 | 154881 | RNAseq | 0.1954 | 0.3602 | |
| SRP133303 | KCTD7 | 154881 | RNAseq | -0.0199 | 0.9244 | |
| SRP159526 | KCTD7 | 154881 | RNAseq | -0.1151 | 0.6128 | |
| SRP193095 | KCTD7 | 154881 | RNAseq | 0.4725 | 0.0002 | |
| SRP219564 | KCTD7 | 154881 | RNAseq | -0.0083 | 0.9869 | |
| TCGA | KCTD7 | 154881 | RNAseq | -0.3109 | 0.0008 |
Upregulated datasets: 0; Downregulated datasets: 0.
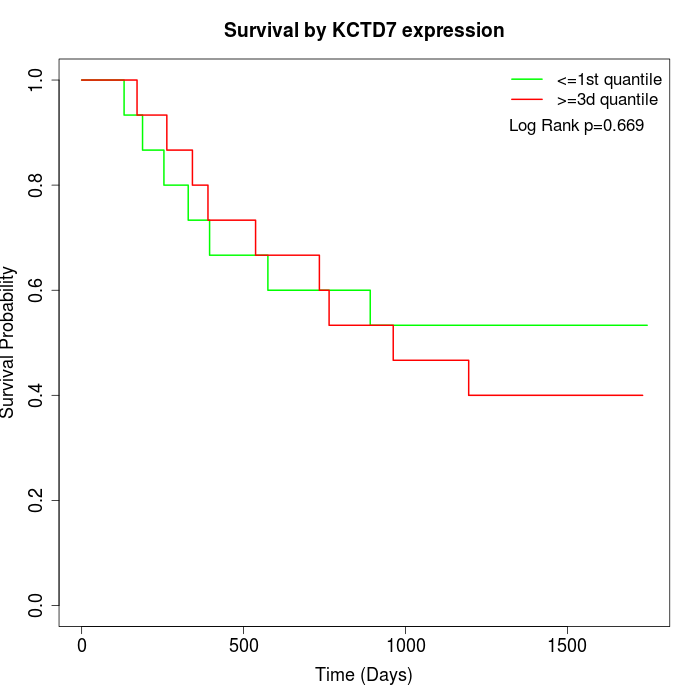
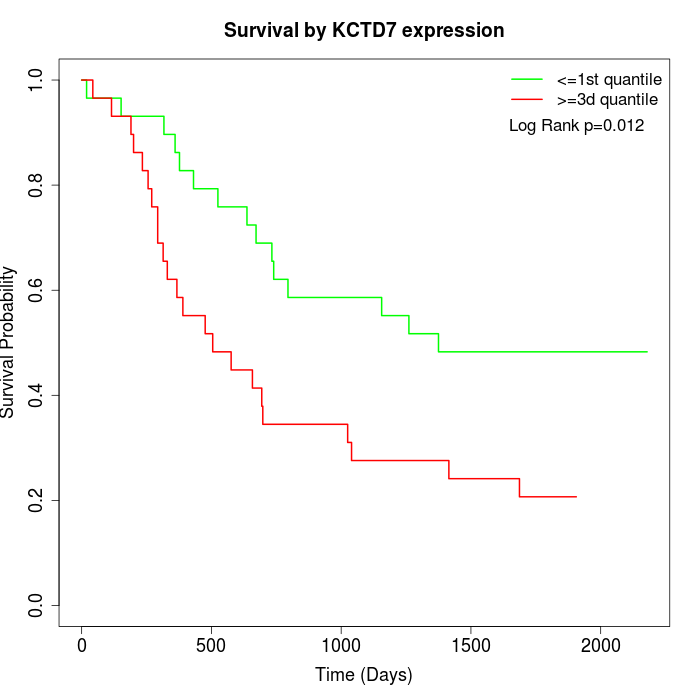
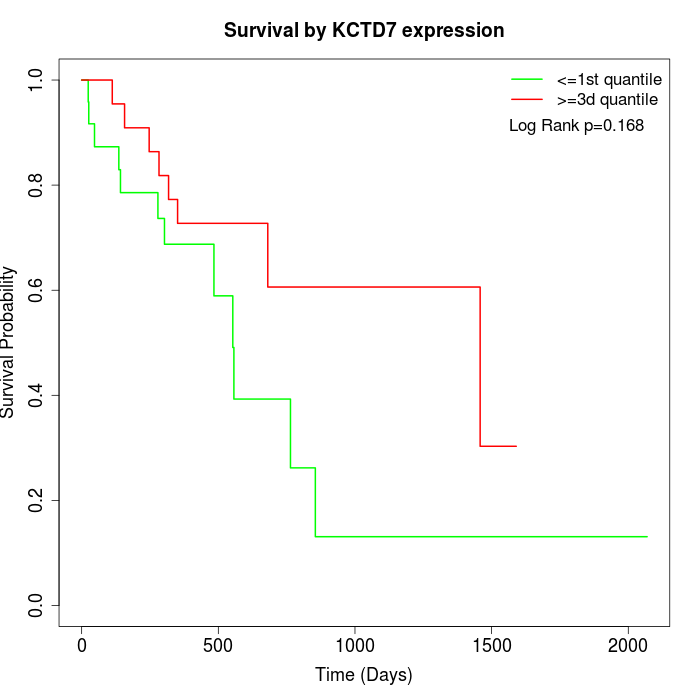
Survival by KCTD7 expression:
Note: Click image to view full size file.
Copy number change of KCTD7:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KCTD7 | 154881 | 11 | 2 | 17 | |
| GSE20123 | KCTD7 | 154881 | 10 | 2 | 18 | |
| GSE43470 | KCTD7 | 154881 | 3 | 1 | 39 | |
| GSE46452 | KCTD7 | 154881 | 13 | 0 | 46 | |
| GSE47630 | KCTD7 | 154881 | 7 | 1 | 32 | |
| GSE54993 | KCTD7 | 154881 | 2 | 7 | 61 | |
| GSE54994 | KCTD7 | 154881 | 12 | 4 | 37 | |
| GSE60625 | KCTD7 | 154881 | 0 | 0 | 11 | |
| GSE74703 | KCTD7 | 154881 | 3 | 1 | 32 | |
| GSE74704 | KCTD7 | 154881 | 5 | 1 | 14 | |
| TCGA | KCTD7 | 154881 | 46 | 7 | 43 |
Total number of gains: 112; Total number of losses: 26; Total Number of normals: 350.
Somatic mutations of KCTD7:
Generating mutation plots.
Highly correlated genes for KCTD7:
Showing top 20/45 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KCTD7 | ABI2 | 0.775158 | 3 | 0 | 3 |
| KCTD7 | PEAK1 | 0.73728 | 3 | 0 | 3 |
| KCTD7 | SMURF2 | 0.737088 | 3 | 0 | 3 |
| KCTD7 | PLEKHA3 | 0.731354 | 3 | 0 | 3 |
| KCTD7 | EIF2AK4 | 0.721873 | 3 | 0 | 3 |
| KCTD7 | MRAS | 0.716498 | 3 | 0 | 3 |
| KCTD7 | PLCL1 | 0.702562 | 3 | 0 | 3 |
| KCTD7 | ADH5 | 0.701629 | 3 | 0 | 3 |
| KCTD7 | DSEL | 0.695194 | 3 | 0 | 3 |
| KCTD7 | RFTN1 | 0.693083 | 3 | 0 | 3 |
| KCTD7 | SESTD1 | 0.689787 | 3 | 0 | 3 |
| KCTD7 | RNASEH2B | 0.689627 | 3 | 0 | 3 |
| KCTD7 | DUSP19 | 0.686728 | 4 | 0 | 4 |
| KCTD7 | EXTL2 | 0.681894 | 3 | 0 | 3 |
| KCTD7 | NR2C1 | 0.680344 | 3 | 0 | 3 |
| KCTD7 | PPP3CB | 0.678936 | 3 | 0 | 3 |
| KCTD7 | ZNF362 | 0.678844 | 3 | 0 | 3 |
| KCTD7 | UBR1 | 0.677256 | 3 | 0 | 3 |
| KCTD7 | FAM13B | 0.676389 | 3 | 0 | 3 |
| KCTD7 | CLIC4 | 0.675508 | 3 | 0 | 3 |
For details and further investigation, click here