| Full name: slingshot protein phosphatase 3 | Alias Symbol: FLJ20515|FLJ10928 | ||
| Type: protein-coding gene | Cytoband: 11q13.2 | ||
| Entrez ID: 54961 | HGNC ID: HGNC:30581 | Ensembl Gene: ENSG00000172830 | OMIM ID: 606780 |
| Drug and gene relationship at DGIdb | |||
SSH3 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04360 | Axon guidance | |
| hsa04810 | Regulation of actin cytoskeleton |
Expression of SSH3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SSH3 | 54961 | 51192_at | -0.0931 | 0.8897 | |
| GSE20347 | SSH3 | 54961 | 51192_at | -0.2422 | 0.1344 | |
| GSE23400 | SSH3 | 54961 | 219241_x_at | -0.3299 | 0.0000 | |
| GSE26886 | SSH3 | 54961 | 51192_at | 0.0229 | 0.9177 | |
| GSE29001 | SSH3 | 54961 | 51192_at | -0.4149 | 0.0485 | |
| GSE38129 | SSH3 | 54961 | 51192_at | -0.3589 | 0.0038 | |
| GSE45670 | SSH3 | 54961 | 51192_at | -0.0365 | 0.8361 | |
| GSE53622 | SSH3 | 54961 | 15779 | -0.3138 | 0.0056 | |
| GSE53624 | SSH3 | 54961 | 117394 | -0.7817 | 0.0000 | |
| GSE63941 | SSH3 | 54961 | 51192_at | 0.8230 | 0.1850 | |
| GSE77861 | SSH3 | 54961 | 51192_at | -0.0376 | 0.8841 | |
| GSE97050 | SSH3 | 54961 | A_23_P150147 | 0.0901 | 0.6997 | |
| SRP007169 | SSH3 | 54961 | RNAseq | -1.7016 | 0.0000 | |
| SRP008496 | SSH3 | 54961 | RNAseq | -1.5545 | 0.0000 | |
| SRP064894 | SSH3 | 54961 | RNAseq | -0.6911 | 0.0001 | |
| SRP133303 | SSH3 | 54961 | RNAseq | -0.4124 | 0.0872 | |
| SRP159526 | SSH3 | 54961 | RNAseq | -0.9445 | 0.0026 | |
| SRP193095 | SSH3 | 54961 | RNAseq | -0.3341 | 0.0147 | |
| SRP219564 | SSH3 | 54961 | RNAseq | -0.7000 | 0.0450 | |
| TCGA | SSH3 | 54961 | RNAseq | 0.0121 | 0.8627 |
Upregulated datasets: 0; Downregulated datasets: 2.
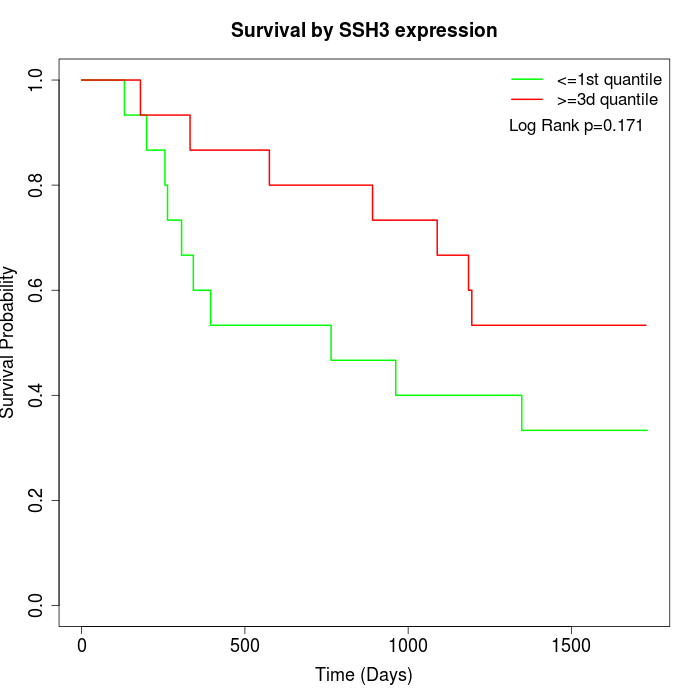
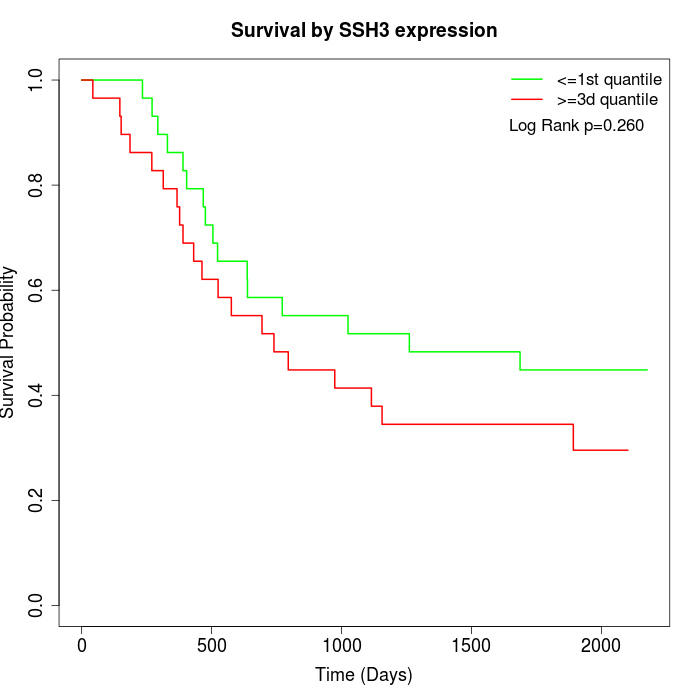
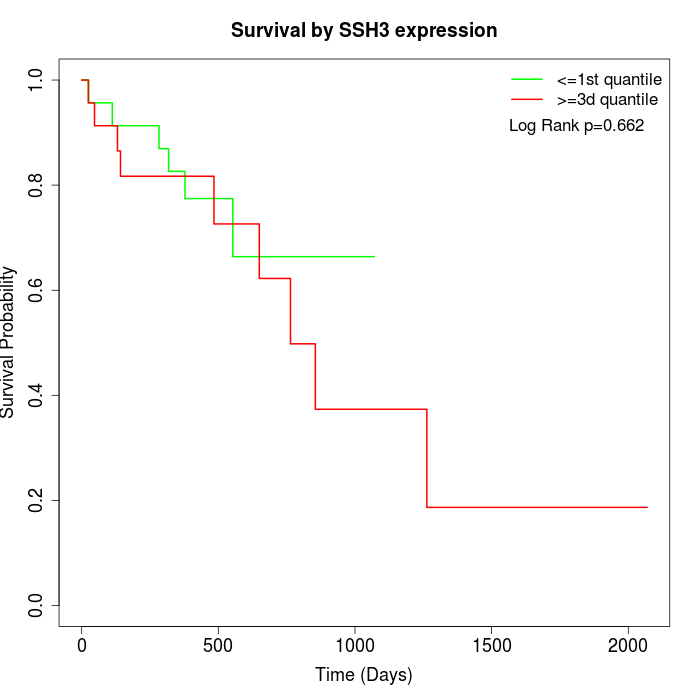
Survival by SSH3 expression:
Note: Click image to view full size file.
Copy number change of SSH3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SSH3 | 54961 | 8 | 4 | 18 | |
| GSE20123 | SSH3 | 54961 | 8 | 4 | 18 | |
| GSE43470 | SSH3 | 54961 | 6 | 1 | 36 | |
| GSE46452 | SSH3 | 54961 | 13 | 3 | 43 | |
| GSE47630 | SSH3 | 54961 | 8 | 5 | 27 | |
| GSE54993 | SSH3 | 54961 | 3 | 0 | 67 | |
| GSE54994 | SSH3 | 54961 | 13 | 4 | 36 | |
| GSE60625 | SSH3 | 54961 | 0 | 3 | 8 | |
| GSE74703 | SSH3 | 54961 | 4 | 0 | 32 | |
| GSE74704 | SSH3 | 54961 | 6 | 2 | 12 | |
| TCGA | SSH3 | 54961 | 28 | 8 | 60 |
Total number of gains: 97; Total number of losses: 34; Total Number of normals: 357.
Somatic mutations of SSH3:
Generating mutation plots.
Highly correlated genes for SSH3:
Showing top 20/371 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SSH3 | TAOK2 | 0.784897 | 4 | 0 | 4 |
| SSH3 | SP8 | 0.736376 | 3 | 0 | 3 |
| SSH3 | LIMD1 | 0.708431 | 4 | 0 | 4 |
| SSH3 | CNGB1 | 0.686998 | 5 | 0 | 4 |
| SSH3 | FBXO25 | 0.679208 | 3 | 0 | 3 |
| SSH3 | KRTAP9-9 | 0.678712 | 3 | 0 | 3 |
| SSH3 | S100A5 | 0.67748 | 4 | 0 | 3 |
| SSH3 | SEMA6C | 0.673856 | 4 | 0 | 4 |
| SSH3 | DNAH6 | 0.665852 | 4 | 0 | 3 |
| SSH3 | TMEM102 | 0.664668 | 4 | 0 | 3 |
| SSH3 | CABP7 | 0.663694 | 3 | 0 | 3 |
| SSH3 | BMP15 | 0.662972 | 3 | 0 | 3 |
| SSH3 | SLC22A8 | 0.66268 | 4 | 0 | 3 |
| SSH3 | HMGB4 | 0.66012 | 4 | 0 | 4 |
| SSH3 | GPR182 | 0.659081 | 4 | 0 | 3 |
| SSH3 | MAST4 | 0.654863 | 10 | 0 | 9 |
| SSH3 | EFNA2 | 0.652201 | 4 | 0 | 3 |
| SSH3 | CABP1 | 0.651399 | 4 | 0 | 3 |
| SSH3 | FUZ | 0.649026 | 4 | 0 | 3 |
| SSH3 | NPAS1 | 0.648586 | 4 | 0 | 3 |
For details and further investigation, click here