| Full name: keratin associated protein 9-9 | Alias Symbol: KAP9.9|KAP9.5 | ||
| Type: protein-coding gene | Cytoband: 17q21.2 | ||
| Entrez ID: 81870 | HGNC ID: HGNC:16773 | Ensembl Gene: ENSG00000198083 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of KRTAP9-9:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | KRTAP9-9 | 81870 | 220972_s_at | 0.0093 | 0.9779 | |
| GSE20347 | KRTAP9-9 | 81870 | 220972_s_at | -0.0151 | 0.7480 | |
| GSE23400 | KRTAP9-9 | 81870 | 220972_s_at | -0.0656 | 0.0064 | |
| GSE26886 | KRTAP9-9 | 81870 | 220972_s_at | 0.0402 | 0.7026 | |
| GSE29001 | KRTAP9-9 | 81870 | 220972_s_at | -0.0559 | 0.6383 | |
| GSE38129 | KRTAP9-9 | 81870 | 220972_s_at | -0.0279 | 0.5292 | |
| GSE45670 | KRTAP9-9 | 81870 | 220972_s_at | 0.1261 | 0.0574 | |
| GSE53622 | KRTAP9-9 | 81870 | 98407 | 0.0915 | 0.5538 | |
| GSE53624 | KRTAP9-9 | 81870 | 98407 | 0.2252 | 0.0097 | |
| GSE63941 | KRTAP9-9 | 81870 | 220972_s_at | 0.0710 | 0.4711 | |
| GSE77861 | KRTAP9-9 | 81870 | 220972_s_at | -0.0587 | 0.4061 | |
| GSE97050 | KRTAP9-9 | 81870 | A_23_P311585 | 0.0229 | 0.9236 |
Upregulated datasets: 0; Downregulated datasets: 0.
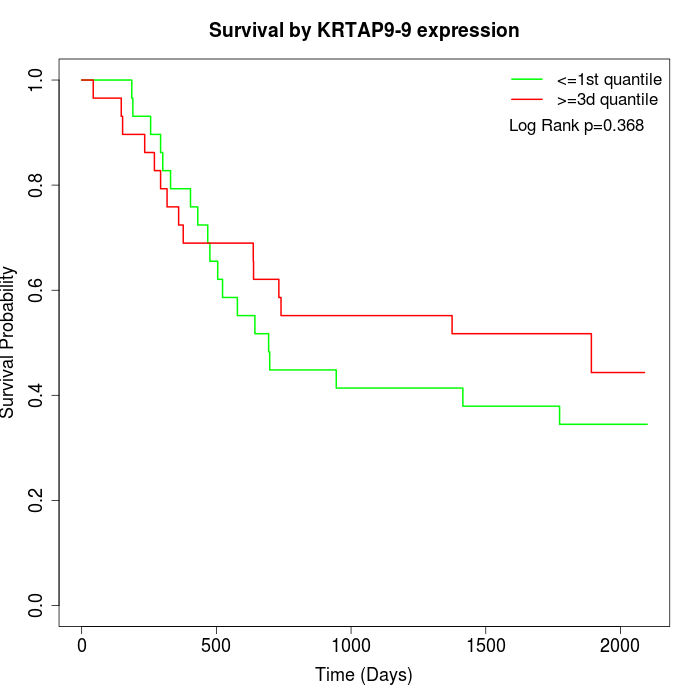
Survival by KRTAP9-9 expression:
Note: Click image to view full size file.
Copy number change of KRTAP9-9:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | KRTAP9-9 | 81870 | 6 | 2 | 22 | |
| GSE20123 | KRTAP9-9 | 81870 | 6 | 2 | 22 | |
| GSE43470 | KRTAP9-9 | 81870 | 1 | 2 | 40 | |
| GSE46452 | KRTAP9-9 | 81870 | 34 | 0 | 25 | |
| GSE47630 | KRTAP9-9 | 81870 | 8 | 1 | 31 | |
| GSE54993 | KRTAP9-9 | 81870 | 3 | 4 | 63 | |
| GSE54994 | KRTAP9-9 | 81870 | 8 | 5 | 40 | |
| GSE60625 | KRTAP9-9 | 81870 | 4 | 0 | 7 | |
| GSE74703 | KRTAP9-9 | 81870 | 1 | 1 | 34 | |
| GSE74704 | KRTAP9-9 | 81870 | 4 | 1 | 15 |
Total number of gains: 75; Total number of losses: 18; Total Number of normals: 299.
Somatic mutations of KRTAP9-9:
Generating mutation plots.
Highly correlated genes for KRTAP9-9:
Showing top 20/294 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| KRTAP9-9 | C3orf36 | 0.78959 | 3 | 0 | 3 |
| KRTAP9-9 | KRT35 | 0.746414 | 3 | 0 | 3 |
| KRTAP9-9 | SLC9A7 | 0.741468 | 3 | 0 | 3 |
| KRTAP9-9 | PRAMEF12 | 0.740088 | 3 | 0 | 3 |
| KRTAP9-9 | CACNA1I | 0.730426 | 3 | 0 | 3 |
| KRTAP9-9 | PTGDR2 | 0.729372 | 3 | 0 | 3 |
| KRTAP9-9 | ZNF81 | 0.720633 | 3 | 0 | 3 |
| KRTAP9-9 | SI | 0.714011 | 3 | 0 | 3 |
| KRTAP9-9 | RIN1 | 0.712645 | 3 | 0 | 3 |
| KRTAP9-9 | TUBA8 | 0.709879 | 3 | 0 | 3 |
| KRTAP9-9 | PKLR | 0.703981 | 4 | 0 | 3 |
| KRTAP9-9 | NCR3 | 0.693063 | 4 | 0 | 3 |
| KRTAP9-9 | ALAS2 | 0.687054 | 3 | 0 | 3 |
| KRTAP9-9 | GRK4 | 0.684467 | 3 | 0 | 3 |
| KRTAP9-9 | CYP11B1 | 0.68318 | 4 | 0 | 3 |
| KRTAP9-9 | FGF5 | 0.682923 | 4 | 0 | 4 |
| KRTAP9-9 | TMEM59L | 0.681 | 3 | 0 | 3 |
| KRTAP9-9 | KCNC1 | 0.679465 | 3 | 0 | 3 |
| KRTAP9-9 | SSH3 | 0.678712 | 3 | 0 | 3 |
| KRTAP9-9 | FASLG | 0.677649 | 5 | 0 | 4 |
For details and further investigation, click here