| Full name: T-box brain transcription factor 1 | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 2q24.2 | ||
| Entrez ID: 10716 | HGNC ID: HGNC:11590 | Ensembl Gene: ENSG00000136535 | OMIM ID: 604616 |
| Drug and gene relationship at DGIdb | |||
Expression of TBR1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TBR1 | 10716 | 220025_at | -0.0715 | 0.7716 | |
| GSE20347 | TBR1 | 10716 | 220025_at | -0.0330 | 0.6021 | |
| GSE23400 | TBR1 | 10716 | 220025_at | 0.0164 | 0.6608 | |
| GSE26886 | TBR1 | 10716 | 220025_at | 0.0724 | 0.4178 | |
| GSE29001 | TBR1 | 10716 | 220025_at | -0.1107 | 0.6734 | |
| GSE38129 | TBR1 | 10716 | 220025_at | -0.0483 | 0.4087 | |
| GSE45670 | TBR1 | 10716 | 220025_at | 0.0127 | 0.8691 | |
| GSE53622 | TBR1 | 10716 | 138342 | 0.2392 | 0.0025 | |
| GSE53624 | TBR1 | 10716 | 138342 | 0.3543 | 0.0000 | |
| GSE63941 | TBR1 | 10716 | 220025_at | 0.0255 | 0.8501 | |
| GSE77861 | TBR1 | 10716 | 220025_at | 0.0063 | 0.9563 | |
| TCGA | TBR1 | 10716 | RNAseq | 2.4945 | 0.0223 |
Upregulated datasets: 1; Downregulated datasets: 0.
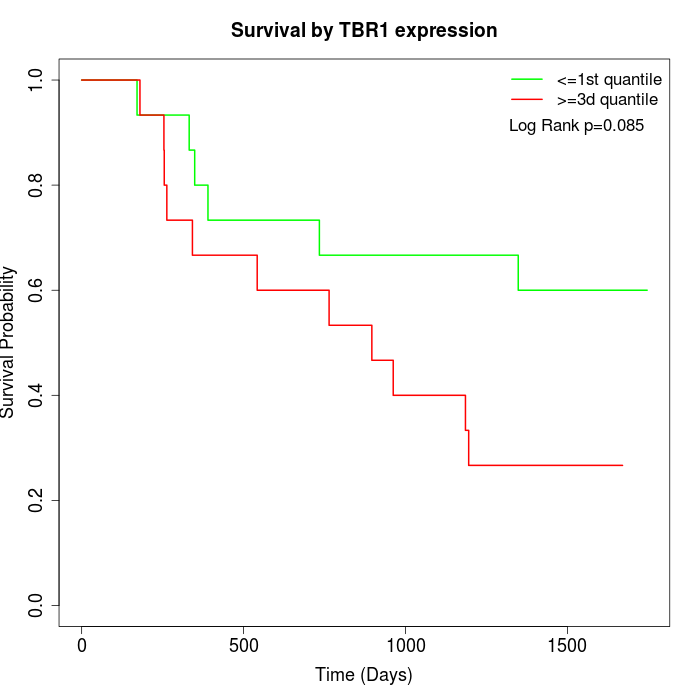
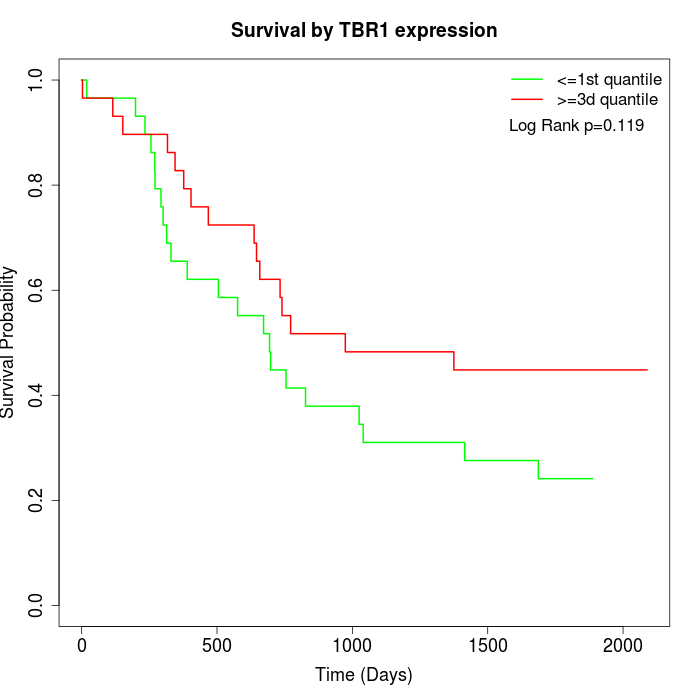
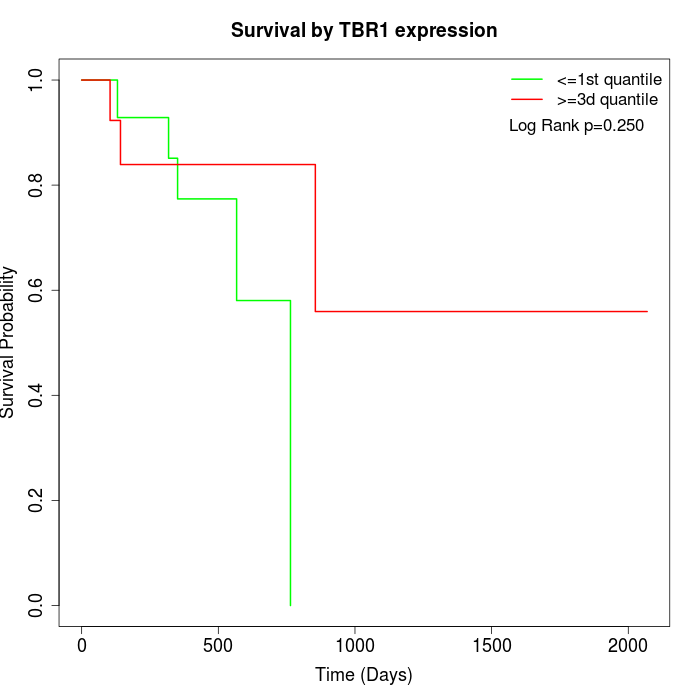
Survival by TBR1 expression:
Note: Click image to view full size file.
Copy number change of TBR1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TBR1 | 10716 | 4 | 1 | 25 | |
| GSE20123 | TBR1 | 10716 | 4 | 1 | 25 | |
| GSE43470 | TBR1 | 10716 | 4 | 1 | 38 | |
| GSE46452 | TBR1 | 10716 | 1 | 4 | 54 | |
| GSE47630 | TBR1 | 10716 | 5 | 3 | 32 | |
| GSE54993 | TBR1 | 10716 | 0 | 5 | 65 | |
| GSE54994 | TBR1 | 10716 | 11 | 2 | 40 | |
| GSE60625 | TBR1 | 10716 | 0 | 3 | 8 | |
| GSE74703 | TBR1 | 10716 | 3 | 1 | 32 | |
| GSE74704 | TBR1 | 10716 | 3 | 0 | 17 | |
| TCGA | TBR1 | 10716 | 23 | 10 | 63 |
Total number of gains: 58; Total number of losses: 31; Total Number of normals: 399.
Somatic mutations of TBR1:
Generating mutation plots.
Highly correlated genes for TBR1:
Showing top 20/449 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TBR1 | TEX15 | 0.705077 | 3 | 0 | 3 |
| TBR1 | STXBP5L | 0.691383 | 3 | 0 | 3 |
| TBR1 | CER1 | 0.687799 | 3 | 0 | 3 |
| TBR1 | KCNMB2 | 0.686745 | 4 | 0 | 3 |
| TBR1 | GPR22 | 0.684373 | 4 | 0 | 3 |
| TBR1 | CHST5 | 0.681018 | 3 | 0 | 3 |
| TBR1 | HTR1D | 0.680852 | 4 | 0 | 3 |
| TBR1 | CLCN4 | 0.68044 | 3 | 0 | 3 |
| TBR1 | HAVCR1 | 0.680058 | 3 | 0 | 3 |
| TBR1 | NHLH1 | 0.678237 | 5 | 0 | 5 |
| TBR1 | EDDM3A | 0.678175 | 4 | 0 | 3 |
| TBR1 | HNF1B | 0.675798 | 4 | 0 | 3 |
| TBR1 | C2orf83 | 0.671318 | 3 | 0 | 3 |
| TBR1 | PRPS1L1 | 0.671148 | 5 | 0 | 5 |
| TBR1 | RPS6KA6 | 0.668056 | 5 | 0 | 4 |
| TBR1 | HGH1 | 0.665614 | 3 | 0 | 3 |
| TBR1 | OR10C1 | 0.660177 | 5 | 0 | 4 |
| TBR1 | MIER2 | 0.658974 | 4 | 0 | 3 |
| TBR1 | CCL25 | 0.658402 | 4 | 0 | 4 |
| TBR1 | SLC17A3 | 0.655941 | 3 | 0 | 3 |
For details and further investigation, click here