| Full name: transglutaminase 2 | Alias Symbol: TGC | ||
| Type: protein-coding gene | Cytoband: 20q11.23 | ||
| Entrez ID: 7052 | HGNC ID: HGNC:11778 | Ensembl Gene: ENSG00000198959 | OMIM ID: 190196 |
| Drug and gene relationship at DGIdb | |||
TGM2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa05016 | Huntington's disease |
Expression of TGM2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TGM2 | 7052 | 201042_at | -0.5861 | 0.5751 | |
| GSE20347 | TGM2 | 7052 | 201042_at | 0.0310 | 0.8900 | |
| GSE23400 | TGM2 | 7052 | 201042_at | 0.0652 | 0.6541 | |
| GSE26886 | TGM2 | 7052 | 201042_at | 1.0488 | 0.0107 | |
| GSE29001 | TGM2 | 7052 | 201042_at | 0.2866 | 0.5506 | |
| GSE38129 | TGM2 | 7052 | 201042_at | -0.2664 | 0.5212 | |
| GSE45670 | TGM2 | 7052 | 201042_at | -0.7843 | 0.0222 | |
| GSE53622 | TGM2 | 7052 | 301 | -0.1929 | 0.2371 | |
| GSE53624 | TGM2 | 7052 | 301 | 0.0530 | 0.7132 | |
| GSE63941 | TGM2 | 7052 | 201042_at | -2.3554 | 0.0232 | |
| GSE77861 | TGM2 | 7052 | 211573_x_at | 0.0140 | 0.9353 | |
| GSE97050 | TGM2 | 7052 | A_32_P86763 | 0.4441 | 0.2302 | |
| SRP007169 | TGM2 | 7052 | RNAseq | 4.0461 | 0.0000 | |
| SRP008496 | TGM2 | 7052 | RNAseq | 4.3938 | 0.0000 | |
| SRP064894 | TGM2 | 7052 | RNAseq | 0.9123 | 0.0223 | |
| SRP133303 | TGM2 | 7052 | RNAseq | 0.6131 | 0.2202 | |
| SRP159526 | TGM2 | 7052 | RNAseq | 0.4343 | 0.5068 | |
| SRP193095 | TGM2 | 7052 | RNAseq | 2.0996 | 0.0000 | |
| SRP219564 | TGM2 | 7052 | RNAseq | 0.6412 | 0.4420 | |
| TCGA | TGM2 | 7052 | RNAseq | -0.2615 | 0.0385 |
Upregulated datasets: 4; Downregulated datasets: 1.
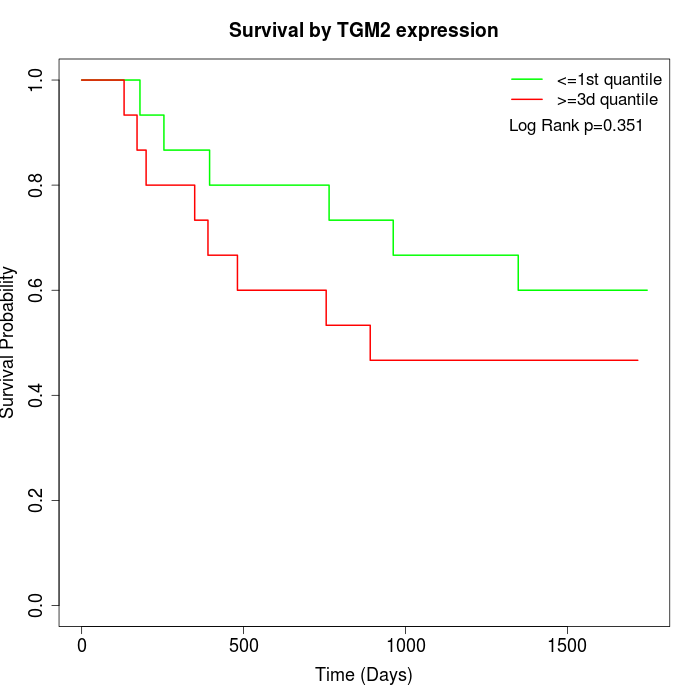
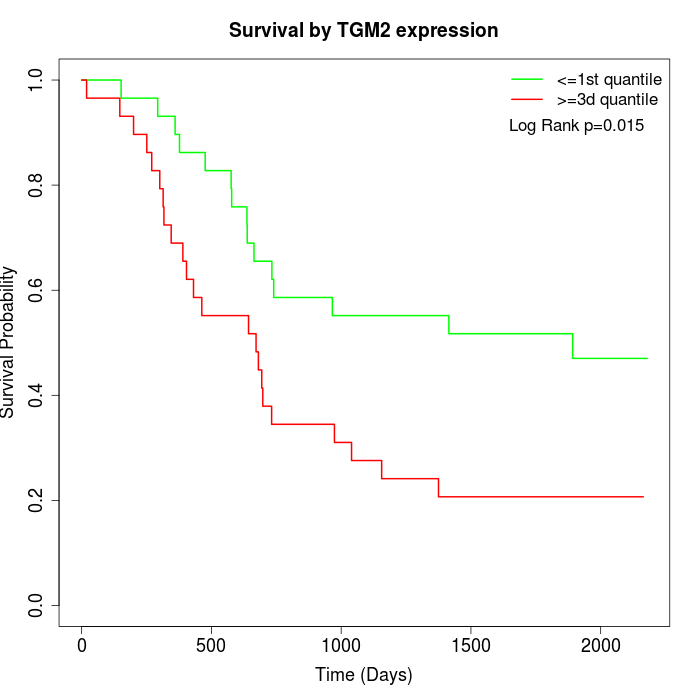
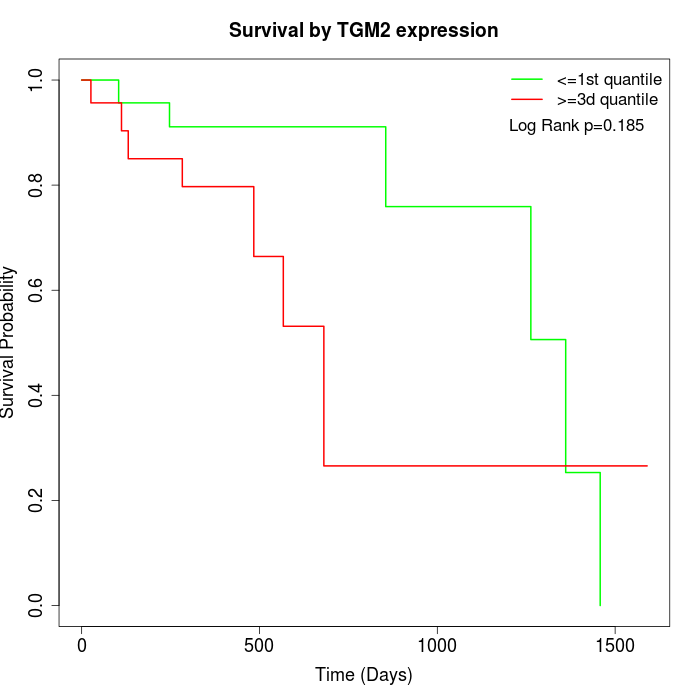
Survival by TGM2 expression:
Note: Click image to view full size file.
Copy number change of TGM2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TGM2 | 7052 | 14 | 2 | 14 | |
| GSE20123 | TGM2 | 7052 | 14 | 2 | 14 | |
| GSE43470 | TGM2 | 7052 | 12 | 0 | 31 | |
| GSE46452 | TGM2 | 7052 | 29 | 0 | 30 | |
| GSE47630 | TGM2 | 7052 | 24 | 0 | 16 | |
| GSE54993 | TGM2 | 7052 | 0 | 17 | 53 | |
| GSE54994 | TGM2 | 7052 | 23 | 1 | 29 | |
| GSE60625 | TGM2 | 7052 | 0 | 0 | 11 | |
| GSE74703 | TGM2 | 7052 | 10 | 0 | 26 | |
| GSE74704 | TGM2 | 7052 | 10 | 1 | 9 | |
| TCGA | TGM2 | 7052 | 47 | 4 | 45 |
Total number of gains: 183; Total number of losses: 27; Total Number of normals: 278.
Somatic mutations of TGM2:
Generating mutation plots.
Highly correlated genes for TGM2:
Showing top 20/1091 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TGM2 | THYN1 | 0.776712 | 3 | 0 | 3 |
| TGM2 | HAS1 | 0.76299 | 3 | 0 | 3 |
| TGM2 | C11orf96 | 0.741067 | 5 | 0 | 5 |
| TGM2 | TNFRSF19 | 0.740486 | 3 | 0 | 3 |
| TGM2 | RBMX2 | 0.735254 | 3 | 0 | 3 |
| TGM2 | CNTLN | 0.733866 | 3 | 0 | 3 |
| TGM2 | UBXN1 | 0.729015 | 3 | 0 | 3 |
| TGM2 | LETM2 | 0.727114 | 3 | 0 | 3 |
| TGM2 | RFTN1 | 0.725734 | 9 | 0 | 9 |
| TGM2 | FHL3 | 0.720533 | 3 | 0 | 3 |
| TGM2 | ANXA6 | 0.71985 | 10 | 0 | 9 |
| TGM2 | NMT2 | 0.716628 | 4 | 0 | 4 |
| TGM2 | GLIS3 | 0.715701 | 4 | 0 | 4 |
| TGM2 | A1BG | 0.715345 | 3 | 0 | 3 |
| TGM2 | ING1 | 0.706596 | 3 | 0 | 3 |
| TGM2 | KPNA6 | 0.706536 | 3 | 0 | 3 |
| TGM2 | PLEKHO1 | 0.705549 | 12 | 0 | 11 |
| TGM2 | KCNJ8 | 0.704151 | 11 | 0 | 11 |
| TGM2 | TGFB1I1 | 0.703051 | 10 | 0 | 10 |
| TGM2 | MCAM | 0.702503 | 10 | 0 | 9 |
For details and further investigation, click here