| Full name: enoyl-CoA hydratase, short chain 1 | Alias Symbol: SCEH | ||
| Type: protein-coding gene | Cytoband: 10q26.3 | ||
| Entrez ID: 1892 | HGNC ID: HGNC:3151 | Ensembl Gene: ENSG00000127884 | OMIM ID: 602292 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of ECHS1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ECHS1 | 1892 | 201135_at | -0.1331 | 0.8196 | |
| GSE20347 | ECHS1 | 1892 | 201135_at | -0.3883 | 0.0155 | |
| GSE23400 | ECHS1 | 1892 | 201135_at | -0.4374 | 0.0000 | |
| GSE26886 | ECHS1 | 1892 | 201135_at | -0.7372 | 0.0001 | |
| GSE29001 | ECHS1 | 1892 | 201135_at | -0.6053 | 0.0295 | |
| GSE38129 | ECHS1 | 1892 | 201135_at | -0.3729 | 0.0074 | |
| GSE45670 | ECHS1 | 1892 | 201135_at | -0.3130 | 0.0159 | |
| GSE53622 | ECHS1 | 1892 | 12113 | -0.7313 | 0.0000 | |
| GSE53624 | ECHS1 | 1892 | 12113 | -0.6384 | 0.0000 | |
| GSE63941 | ECHS1 | 1892 | 201135_at | -0.0910 | 0.8440 | |
| GSE77861 | ECHS1 | 1892 | 201135_at | -0.7799 | 0.0094 | |
| GSE97050 | ECHS1 | 1892 | A_23_P104362 | -0.2901 | 0.2349 | |
| SRP007169 | ECHS1 | 1892 | RNAseq | -1.1057 | 0.0004 | |
| SRP008496 | ECHS1 | 1892 | RNAseq | -1.1426 | 0.0000 | |
| SRP064894 | ECHS1 | 1892 | RNAseq | -0.4434 | 0.1290 | |
| SRP133303 | ECHS1 | 1892 | RNAseq | -0.4397 | 0.0015 | |
| SRP159526 | ECHS1 | 1892 | RNAseq | -0.5569 | 0.0248 | |
| SRP193095 | ECHS1 | 1892 | RNAseq | -0.7769 | 0.0000 | |
| SRP219564 | ECHS1 | 1892 | RNAseq | -0.5039 | 0.0992 | |
| TCGA | ECHS1 | 1892 | RNAseq | -0.1454 | 0.0052 |
Upregulated datasets: 0; Downregulated datasets: 2.
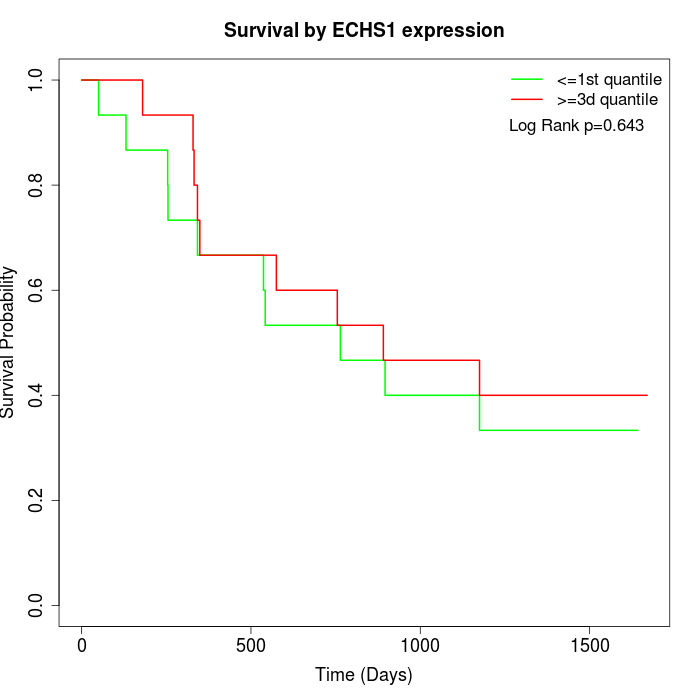
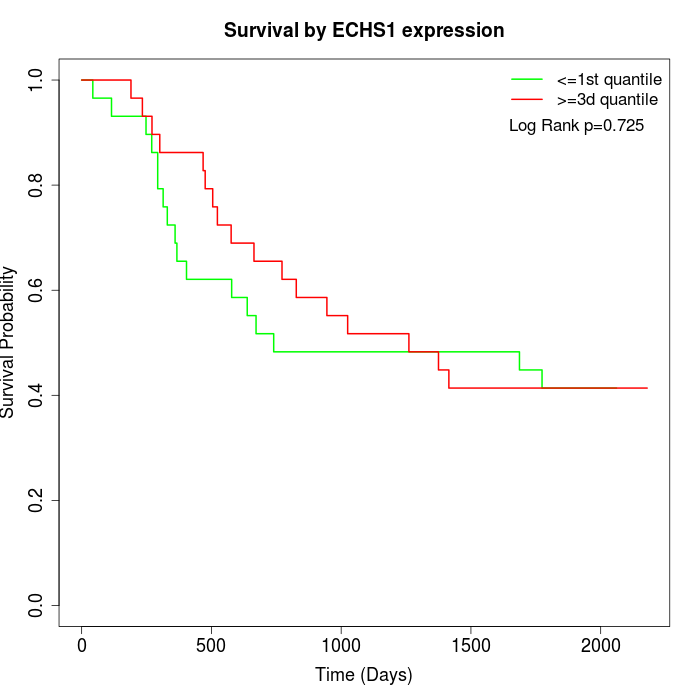
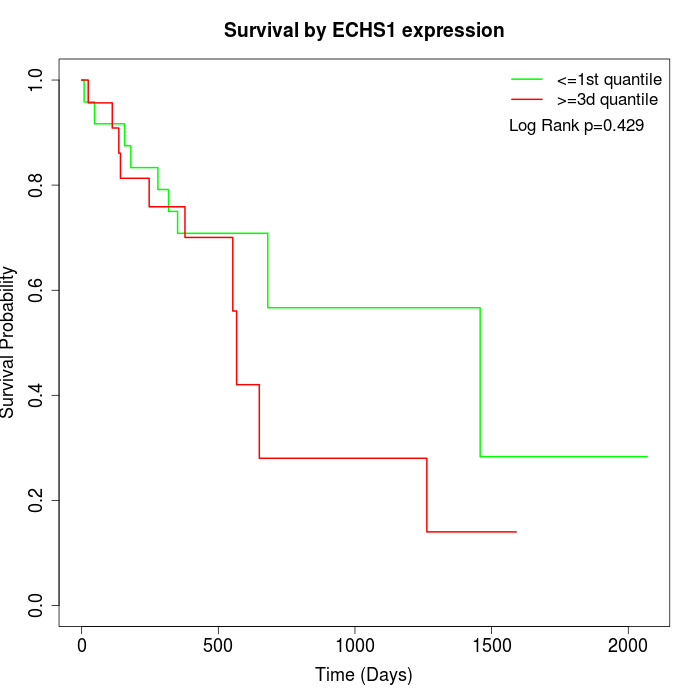
Survival by ECHS1 expression:
Note: Click image to view full size file.
Copy number change of ECHS1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ECHS1 | 1892 | 3 | 8 | 19 | |
| GSE20123 | ECHS1 | 1892 | 3 | 9 | 18 | |
| GSE43470 | ECHS1 | 1892 | 0 | 8 | 35 | |
| GSE46452 | ECHS1 | 1892 | 1 | 11 | 47 | |
| GSE47630 | ECHS1 | 1892 | 4 | 13 | 23 | |
| GSE54993 | ECHS1 | 1892 | 8 | 2 | 60 | |
| GSE54994 | ECHS1 | 1892 | 1 | 9 | 43 | |
| GSE60625 | ECHS1 | 1892 | 0 | 0 | 11 | |
| GSE74703 | ECHS1 | 1892 | 0 | 6 | 30 | |
| GSE74704 | ECHS1 | 1892 | 3 | 4 | 13 | |
| TCGA | ECHS1 | 1892 | 7 | 28 | 61 |
Total number of gains: 30; Total number of losses: 98; Total Number of normals: 360.
Somatic mutations of ECHS1:
Generating mutation plots.
Highly correlated genes for ECHS1:
Showing top 20/1432 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ECHS1 | THAP2 | 0.80354 | 3 | 0 | 3 |
| ECHS1 | ARL16 | 0.740193 | 3 | 0 | 3 |
| ECHS1 | CCDC12 | 0.722056 | 6 | 0 | 6 |
| ECHS1 | PITHD1 | 0.720955 | 7 | 0 | 6 |
| ECHS1 | CA13 | 0.713287 | 3 | 0 | 3 |
| ECHS1 | PEPD | 0.712579 | 4 | 0 | 3 |
| ECHS1 | CCDC6 | 0.711111 | 11 | 0 | 11 |
| ECHS1 | SLC46A2 | 0.708841 | 6 | 0 | 6 |
| ECHS1 | FDXACB1 | 0.706921 | 3 | 0 | 3 |
| ECHS1 | VCPIP1 | 0.70474 | 3 | 0 | 3 |
| ECHS1 | RFXANK | 0.703755 | 3 | 0 | 3 |
| ECHS1 | TDRG1 | 0.703572 | 3 | 0 | 3 |
| ECHS1 | CHCHD1 | 0.702337 | 7 | 0 | 6 |
| ECHS1 | SRD5A3 | 0.701459 | 3 | 0 | 3 |
| ECHS1 | CETN2 | 0.700539 | 3 | 0 | 3 |
| ECHS1 | ULK3 | 0.697691 | 6 | 0 | 6 |
| ECHS1 | UBE2NL | 0.696769 | 3 | 0 | 3 |
| ECHS1 | ZACN | 0.6965 | 4 | 0 | 4 |
| ECHS1 | USE1 | 0.695352 | 3 | 0 | 3 |
| ECHS1 | RBM20 | 0.694387 | 4 | 0 | 4 |
For details and further investigation, click here