| Full name: translocase of inner mitochondrial membrane 22 | Alias Symbol: TIM22 | ||
| Type: protein-coding gene | Cytoband: 17p13 | ||
| Entrez ID: 29928 | HGNC ID: HGNC:17317 | Ensembl Gene: ENSG00000177370 | OMIM ID: 607251 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of TIMM22:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TIMM22 | 29928 | 219184_x_at | 0.3732 | 0.3997 | |
| GSE20347 | TIMM22 | 29928 | 219184_x_at | 0.2810 | 0.0474 | |
| GSE23400 | TIMM22 | 29928 | 219184_x_at | 0.1006 | 0.1197 | |
| GSE26886 | TIMM22 | 29928 | 219184_x_at | 0.0171 | 0.9553 | |
| GSE29001 | TIMM22 | 29928 | 219184_x_at | 0.3000 | 0.3642 | |
| GSE38129 | TIMM22 | 29928 | 219184_x_at | 0.1373 | 0.2637 | |
| GSE45670 | TIMM22 | 29928 | 229100_s_at | 0.0464 | 0.8121 | |
| GSE53622 | TIMM22 | 29928 | 46641 | 0.0954 | 0.0506 | |
| GSE53624 | TIMM22 | 29928 | 46641 | 0.1983 | 0.0000 | |
| GSE63941 | TIMM22 | 29928 | 219184_x_at | -0.8402 | 0.1164 | |
| GSE77861 | TIMM22 | 29928 | 222841_s_at | 0.1911 | 0.4368 | |
| GSE97050 | TIMM22 | 29928 | A_23_P55468 | 0.0206 | 0.9291 | |
| SRP007169 | TIMM22 | 29928 | RNAseq | 0.8821 | 0.0472 | |
| SRP008496 | TIMM22 | 29928 | RNAseq | 0.7675 | 0.0188 | |
| SRP064894 | TIMM22 | 29928 | RNAseq | 0.6213 | 0.0086 | |
| SRP133303 | TIMM22 | 29928 | RNAseq | -0.0661 | 0.6284 | |
| SRP159526 | TIMM22 | 29928 | RNAseq | 0.4251 | 0.0235 | |
| SRP193095 | TIMM22 | 29928 | RNAseq | 0.1893 | 0.0240 | |
| SRP219564 | TIMM22 | 29928 | RNAseq | 0.2648 | 0.4847 | |
| TCGA | TIMM22 | 29928 | RNAseq | 0.0783 | 0.2105 |
Upregulated datasets: 0; Downregulated datasets: 0.
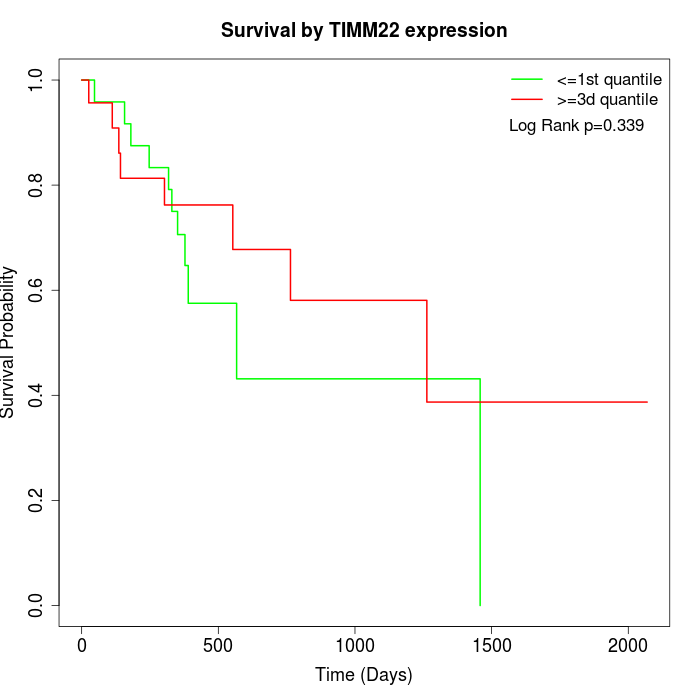
Survival by TIMM22 expression:
Note: Click image to view full size file.
Copy number change of TIMM22:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TIMM22 | 29928 | 5 | 2 | 23 | |
| GSE20123 | TIMM22 | 29928 | 5 | 3 | 22 | |
| GSE43470 | TIMM22 | 29928 | 3 | 6 | 34 | |
| GSE46452 | TIMM22 | 29928 | 34 | 1 | 24 | |
| GSE47630 | TIMM22 | 29928 | 8 | 1 | 31 | |
| GSE54993 | TIMM22 | 29928 | 8 | 3 | 59 | |
| GSE54994 | TIMM22 | 29928 | 7 | 9 | 37 | |
| GSE60625 | TIMM22 | 29928 | 4 | 0 | 7 | |
| GSE74703 | TIMM22 | 29928 | 2 | 3 | 31 | |
| GSE74704 | TIMM22 | 29928 | 3 | 1 | 16 | |
| TCGA | TIMM22 | 29928 | 19 | 21 | 56 |
Total number of gains: 98; Total number of losses: 50; Total Number of normals: 340.
Somatic mutations of TIMM22:
Generating mutation plots.
Highly correlated genes for TIMM22:
Showing top 20/47 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TIMM22 | BTBD3 | 0.720097 | 3 | 0 | 3 |
| TIMM22 | ATP7B | 0.711136 | 3 | 0 | 3 |
| TIMM22 | GPR180 | 0.69729 | 3 | 0 | 3 |
| TIMM22 | TXNL4B | 0.693979 | 3 | 0 | 3 |
| TIMM22 | CDK17 | 0.68879 | 3 | 0 | 3 |
| TIMM22 | PRAF2 | 0.681906 | 4 | 0 | 4 |
| TIMM22 | ZNF777 | 0.681162 | 3 | 0 | 3 |
| TIMM22 | ZNF205 | 0.676364 | 4 | 0 | 3 |
| TIMM22 | C19orf25 | 0.671792 | 3 | 0 | 3 |
| TIMM22 | HR | 0.669097 | 3 | 0 | 3 |
| TIMM22 | PTPRH | 0.660088 | 3 | 0 | 3 |
| TIMM22 | SF3A3 | 0.651067 | 3 | 0 | 3 |
| TIMM22 | TOE1 | 0.649226 | 4 | 0 | 3 |
| TIMM22 | MOK | 0.646814 | 4 | 0 | 3 |
| TIMM22 | PON3 | 0.644412 | 3 | 0 | 3 |
| TIMM22 | GNL1 | 0.639059 | 3 | 0 | 3 |
| TIMM22 | TNNC2 | 0.627819 | 4 | 0 | 3 |
| TIMM22 | RHBDL1 | 0.627099 | 4 | 0 | 3 |
| TIMM22 | PRPF6 | 0.62488 | 4 | 0 | 3 |
| TIMM22 | FAM189B | 0.615222 | 5 | 0 | 4 |
For details and further investigation, click here