| Full name: TPD52 like 3 | Alias Symbol: NYD-SP25|D55 | ||
| Type: protein-coding gene | Cytoband: 9p24.1 | ||
| Entrez ID: 89882 | HGNC ID: HGNC:23382 | Ensembl Gene: ENSG00000170777 | OMIM ID: 617567 |
| Drug and gene relationship at DGIdb | |||
Expression of TPD52L3:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TPD52L3 | 89882 | 231554_at | 0.0184 | 0.9693 | |
| GSE26886 | TPD52L3 | 89882 | 231554_at | -0.2308 | 0.0358 | |
| GSE45670 | TPD52L3 | 89882 | 231554_at | -0.0170 | 0.9073 | |
| GSE53622 | TPD52L3 | 89882 | 31058 | 0.0329 | 0.8112 | |
| GSE53624 | TPD52L3 | 89882 | 31058 | 0.2997 | 0.0120 | |
| GSE63941 | TPD52L3 | 89882 | 231554_at | 0.1650 | 0.4899 | |
| GSE77861 | TPD52L3 | 89882 | 231554_at | 0.1032 | 0.3750 | |
| TCGA | TPD52L3 | 89882 | RNAseq | 1.5956 | 0.4508 |
Upregulated datasets: 0; Downregulated datasets: 0.
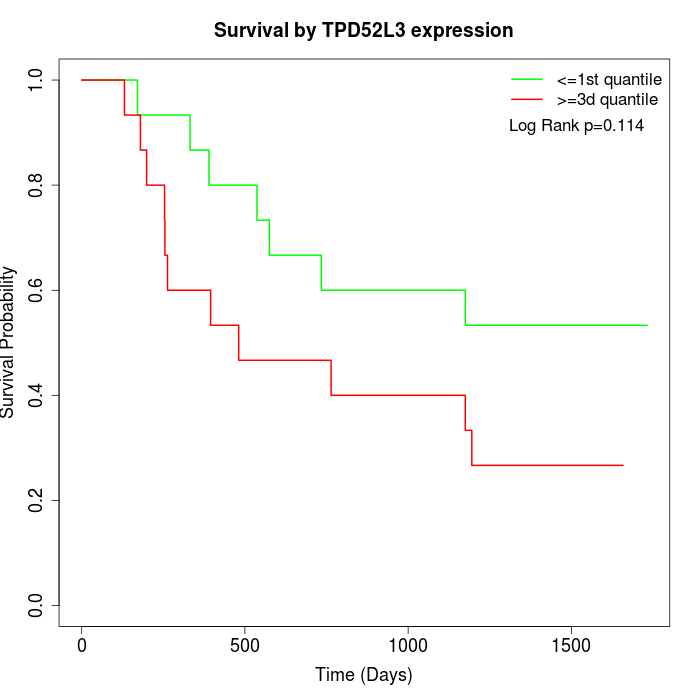
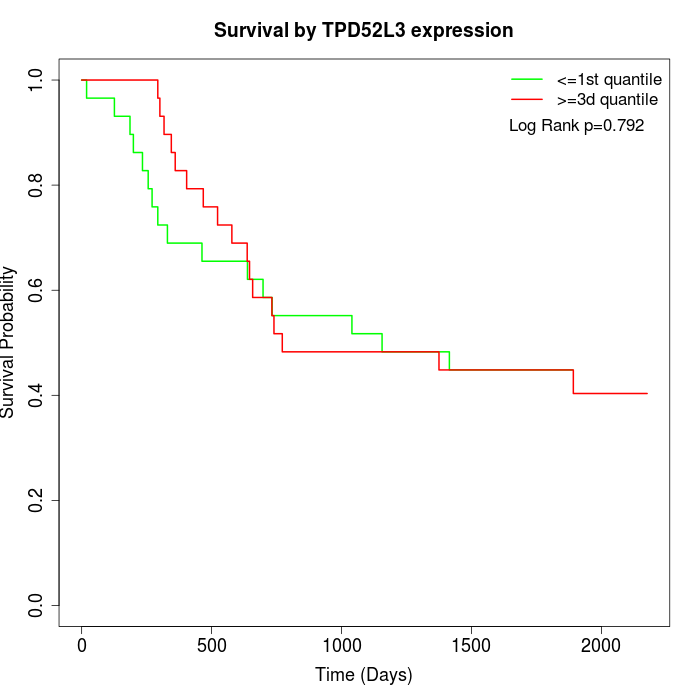
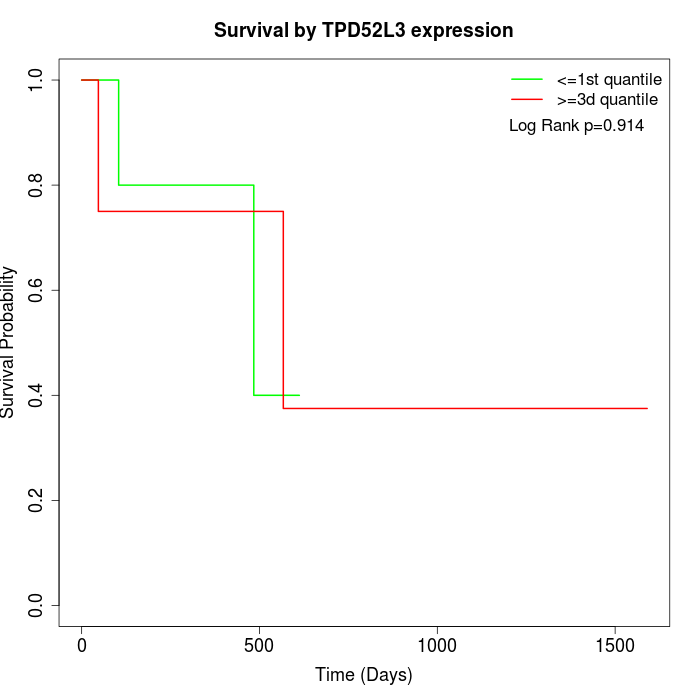
Survival by TPD52L3 expression:
Note: Click image to view full size file.
Copy number change of TPD52L3:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TPD52L3 | 89882 | 1 | 17 | 12 | |
| GSE20123 | TPD52L3 | 89882 | 1 | 17 | 12 | |
| GSE43470 | TPD52L3 | 89882 | 5 | 11 | 27 | |
| GSE46452 | TPD52L3 | 89882 | 2 | 23 | 34 | |
| GSE47630 | TPD52L3 | 89882 | 5 | 26 | 9 | |
| GSE54993 | TPD52L3 | 89882 | 6 | 3 | 61 | |
| GSE54994 | TPD52L3 | 89882 | 9 | 16 | 28 | |
| GSE60625 | TPD52L3 | 89882 | 0 | 2 | 9 | |
| GSE74703 | TPD52L3 | 89882 | 5 | 8 | 23 | |
| GSE74704 | TPD52L3 | 89882 | 0 | 13 | 7 | |
| TCGA | TPD52L3 | 89882 | 14 | 51 | 31 |
Total number of gains: 48; Total number of losses: 187; Total Number of normals: 253.
Somatic mutations of TPD52L3:
Generating mutation plots.
Highly correlated genes for TPD52L3:
Showing top 20/97 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TPD52L3 | ASB17 | 0.697096 | 3 | 0 | 3 |
| TPD52L3 | KIR2DL2 | 0.683875 | 3 | 0 | 3 |
| TPD52L3 | KIR2DS3 | 0.680679 | 4 | 0 | 3 |
| TPD52L3 | TRIM10 | 0.678113 | 3 | 0 | 3 |
| TPD52L3 | ARHGEF38 | 0.668928 | 3 | 0 | 3 |
| TPD52L3 | F2RL3 | 0.654774 | 3 | 0 | 3 |
| TPD52L3 | GSX1 | 0.647742 | 4 | 0 | 3 |
| TPD52L3 | MC2R | 0.636052 | 3 | 0 | 3 |
| TPD52L3 | NRGN | 0.61768 | 3 | 0 | 3 |
| TPD52L3 | CD101 | 0.614352 | 3 | 0 | 3 |
| TPD52L3 | GFRA4 | 0.607302 | 4 | 0 | 3 |
| TPD52L3 | LINC00642 | 0.606443 | 5 | 0 | 4 |
| TPD52L3 | ACACB | 0.599195 | 5 | 0 | 4 |
| TPD52L3 | MAP3K19 | 0.59696 | 5 | 0 | 4 |
| TPD52L3 | SHISA9 | 0.596234 | 3 | 0 | 3 |
| TPD52L3 | KCNQ1OT1 | 0.595324 | 3 | 0 | 3 |
| TPD52L3 | CACNG4 | 0.589599 | 4 | 0 | 3 |
| TPD52L3 | GRIP2 | 0.589147 | 5 | 0 | 3 |
| TPD52L3 | LUZP4 | 0.589013 | 4 | 0 | 3 |
| TPD52L3 | AMBN | 0.58579 | 5 | 0 | 3 |
For details and further investigation, click here