| Full name: basic helix-loop-helix family member e40 | Alias Symbol: DEC1|bHLHe40|SHARP2|Clast5 | ||
| Type: protein-coding gene | Cytoband: 3p26.1 | ||
| Entrez ID: 8553 | HGNC ID: HGNC:1046 | Ensembl Gene: ENSG00000134107 | OMIM ID: 604256 |
| Drug and gene relationship at DGIdb | |||
BHLHE40 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04710 | Circadian rhythm |
Expression of BHLHE40:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BHLHE40 | 8553 | 201170_s_at | -1.4044 | 0.0250 | |
| GSE20347 | BHLHE40 | 8553 | 201170_s_at | -1.2605 | 0.0000 | |
| GSE23400 | BHLHE40 | 8553 | 201170_s_at | -0.6949 | 0.0000 | |
| GSE26886 | BHLHE40 | 8553 | 201170_s_at | -1.3977 | 0.0005 | |
| GSE29001 | BHLHE40 | 8553 | 201170_s_at | -1.3175 | 0.0004 | |
| GSE38129 | BHLHE40 | 8553 | 201170_s_at | -0.8755 | 0.0012 | |
| GSE45670 | BHLHE40 | 8553 | 201170_s_at | -0.7112 | 0.0059 | |
| GSE53622 | BHLHE40 | 8553 | 66465 | -0.9903 | 0.0000 | |
| GSE53624 | BHLHE40 | 8553 | 66465 | -1.0696 | 0.0000 | |
| GSE63941 | BHLHE40 | 8553 | 201170_s_at | 0.9209 | 0.2892 | |
| GSE77861 | BHLHE40 | 8553 | 201170_s_at | -0.3845 | 0.2418 | |
| GSE97050 | BHLHE40 | 8553 | A_24_P268676 | -0.1886 | 0.6139 | |
| SRP007169 | BHLHE40 | 8553 | RNAseq | -1.6538 | 0.0001 | |
| SRP008496 | BHLHE40 | 8553 | RNAseq | -1.7514 | 0.0000 | |
| SRP064894 | BHLHE40 | 8553 | RNAseq | -1.1562 | 0.0003 | |
| SRP133303 | BHLHE40 | 8553 | RNAseq | -0.7344 | 0.0013 | |
| SRP159526 | BHLHE40 | 8553 | RNAseq | -1.9906 | 0.0000 | |
| SRP193095 | BHLHE40 | 8553 | RNAseq | -0.8214 | 0.0003 | |
| SRP219564 | BHLHE40 | 8553 | RNAseq | -0.8516 | 0.1298 | |
| TCGA | BHLHE40 | 8553 | RNAseq | 0.1114 | 0.1249 |
Upregulated datasets: 0; Downregulated datasets: 9.
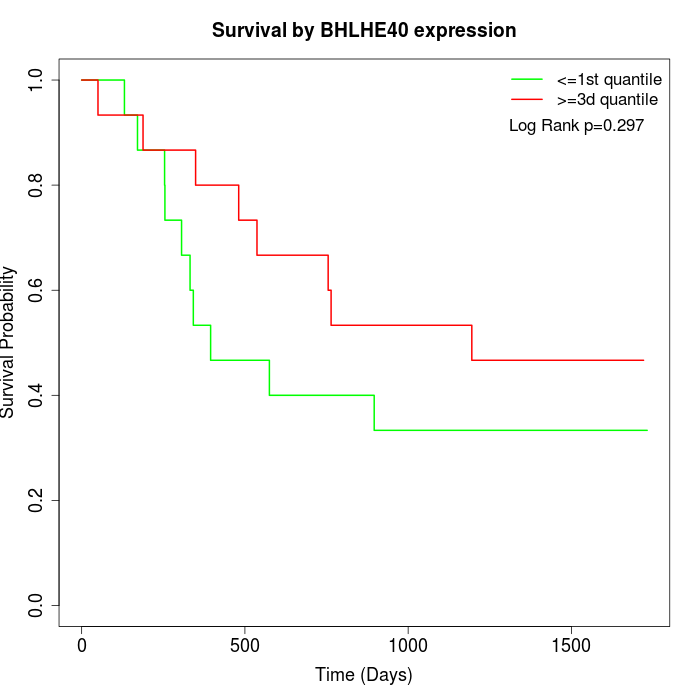
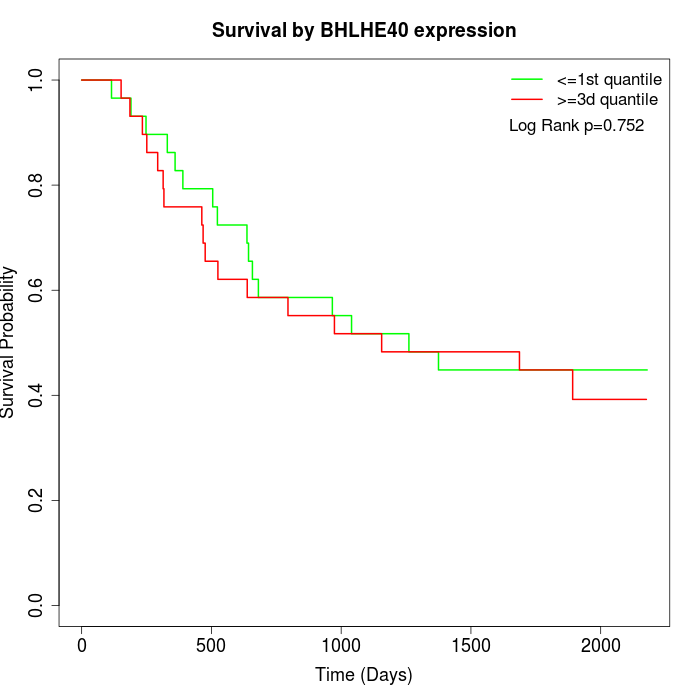
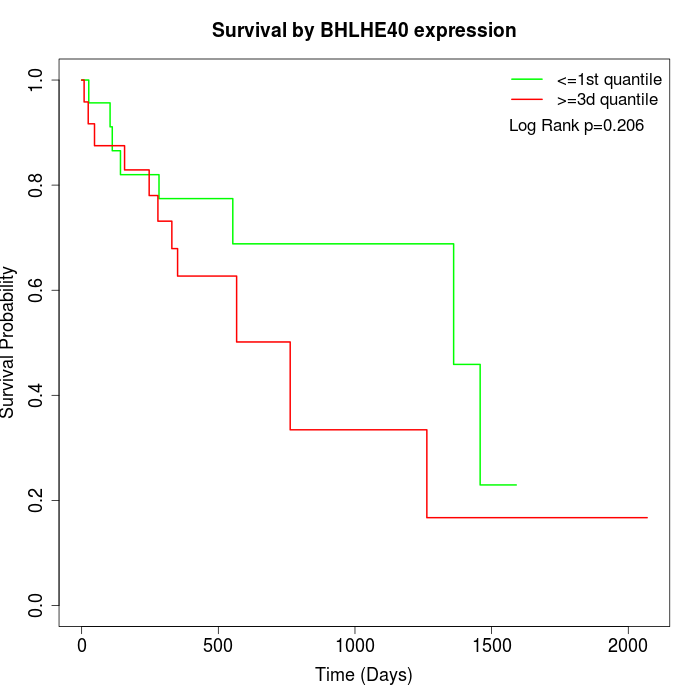
Survival by BHLHE40 expression:
Note: Click image to view full size file.
Copy number change of BHLHE40:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BHLHE40 | 8553 | 1 | 19 | 10 | |
| GSE20123 | BHLHE40 | 8553 | 1 | 19 | 10 | |
| GSE43470 | BHLHE40 | 8553 | 0 | 21 | 22 | |
| GSE46452 | BHLHE40 | 8553 | 2 | 18 | 39 | |
| GSE47630 | BHLHE40 | 8553 | 1 | 24 | 15 | |
| GSE54993 | BHLHE40 | 8553 | 6 | 1 | 63 | |
| GSE54994 | BHLHE40 | 8553 | 1 | 32 | 20 | |
| GSE60625 | BHLHE40 | 8553 | 5 | 0 | 6 | |
| GSE74703 | BHLHE40 | 8553 | 0 | 18 | 18 | |
| GSE74704 | BHLHE40 | 8553 | 1 | 11 | 8 | |
| TCGA | BHLHE40 | 8553 | 1 | 67 | 28 |
Total number of gains: 19; Total number of losses: 230; Total Number of normals: 239.
Somatic mutations of BHLHE40:
Generating mutation plots.
Highly correlated genes for BHLHE40:
Showing top 20/1246 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BHLHE40 | ITPRIP | 0.769799 | 6 | 0 | 6 |
| BHLHE40 | CYSRT1 | 0.734751 | 6 | 0 | 6 |
| BHLHE40 | SERPINB1 | 0.729083 | 10 | 0 | 9 |
| BHLHE40 | TTC37 | 0.725242 | 4 | 0 | 3 |
| BHLHE40 | CNN3 | 0.721296 | 8 | 0 | 8 |
| BHLHE40 | A2ML1 | 0.719772 | 5 | 0 | 5 |
| BHLHE40 | KLF6 | 0.719584 | 11 | 0 | 11 |
| BHLHE40 | CAPN14 | 0.717909 | 5 | 0 | 5 |
| BHLHE40 | SPINK5 | 0.716691 | 9 | 0 | 9 |
| BHLHE40 | EMP1 | 0.712459 | 9 | 0 | 9 |
| BHLHE40 | CLCA4 | 0.710604 | 9 | 0 | 9 |
| BHLHE40 | HIGD1A | 0.709841 | 10 | 0 | 10 |
| BHLHE40 | KRT83 | 0.709462 | 3 | 0 | 3 |
| BHLHE40 | PDCD6IP | 0.707277 | 11 | 0 | 10 |
| BHLHE40 | RHCG | 0.707122 | 9 | 0 | 9 |
| BHLHE40 | SNORA68 | 0.707109 | 3 | 0 | 3 |
| BHLHE40 | TP53I3 | 0.704895 | 10 | 0 | 9 |
| BHLHE40 | PIM1 | 0.7035 | 11 | 0 | 10 |
| BHLHE40 | TMOD3 | 0.703266 | 9 | 0 | 9 |
| BHLHE40 | MPP7 | 0.702356 | 6 | 0 | 6 |
For details and further investigation, click here