| Full name: tubulin alpha 1a | Alias Symbol: TUBA3|B-ALPHA-1|FLJ25113 | ||
| Type: protein-coding gene | Cytoband: 12q13.12 | ||
| Entrez ID: 7846 | HGNC ID: HGNC:20766 | Ensembl Gene: ENSG00000167552 | OMIM ID: 602529 |
| Drug and gene relationship at DGIdb | |||
TUBA1A involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04210 | Apoptosis | |
| hsa05130 | Pathogenic Escherichia coli infection |
Expression of TUBA1A:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TUBA1A | 7846 | 209118_s_at | -0.9277 | 0.3393 | |
| GSE20347 | TUBA1A | 7846 | 209118_s_at | -0.1108 | 0.7310 | |
| GSE23400 | TUBA1A | 7846 | 209118_s_at | -0.0365 | 0.8329 | |
| GSE26886 | TUBA1A | 7846 | 209118_s_at | 0.6019 | 0.0257 | |
| GSE29001 | TUBA1A | 7846 | 209118_s_at | -0.2107 | 0.5385 | |
| GSE38129 | TUBA1A | 7846 | 209118_s_at | -0.1389 | 0.7018 | |
| GSE45670 | TUBA1A | 7846 | 209118_s_at | -0.9655 | 0.0013 | |
| GSE53622 | TUBA1A | 7846 | 87559 | 0.5326 | 0.0000 | |
| GSE53624 | TUBA1A | 7846 | 87559 | 0.5982 | 0.0000 | |
| GSE63941 | TUBA1A | 7846 | 209118_s_at | -3.9561 | 0.0193 | |
| GSE77861 | TUBA1A | 7846 | 209118_s_at | 0.6257 | 0.3039 | |
| GSE97050 | TUBA1A | 7846 | A_23_P139547 | 0.1035 | 0.8234 | |
| SRP007169 | TUBA1A | 7846 | RNAseq | 0.6573 | 0.1054 | |
| SRP008496 | TUBA1A | 7846 | RNAseq | 1.2741 | 0.0002 | |
| SRP064894 | TUBA1A | 7846 | RNAseq | 0.9197 | 0.0171 | |
| SRP133303 | TUBA1A | 7846 | RNAseq | 0.6633 | 0.0212 | |
| SRP159526 | TUBA1A | 7846 | RNAseq | 0.1273 | 0.7437 | |
| SRP193095 | TUBA1A | 7846 | RNAseq | 1.1574 | 0.0000 | |
| SRP219564 | TUBA1A | 7846 | RNAseq | -1.2381 | 0.2351 | |
| TCGA | TUBA1A | 7846 | RNAseq | -0.0450 | 0.5908 |
Upregulated datasets: 2; Downregulated datasets: 1.
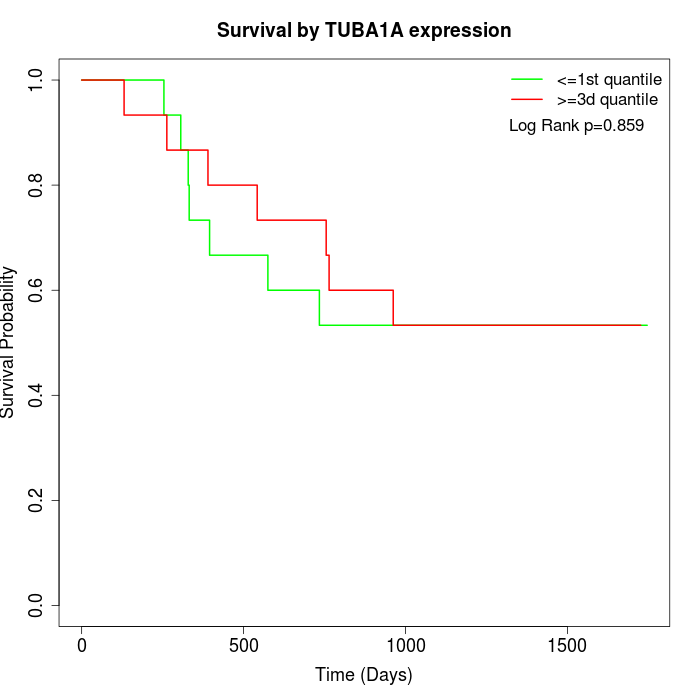
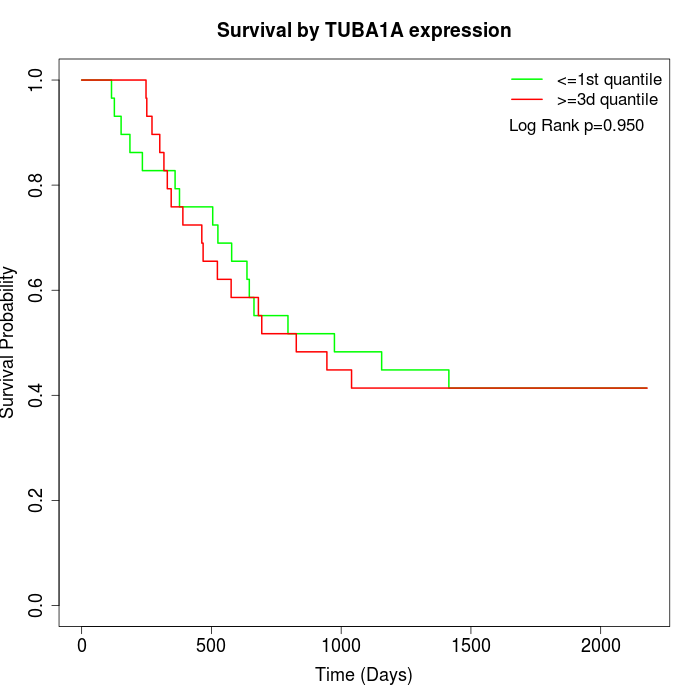
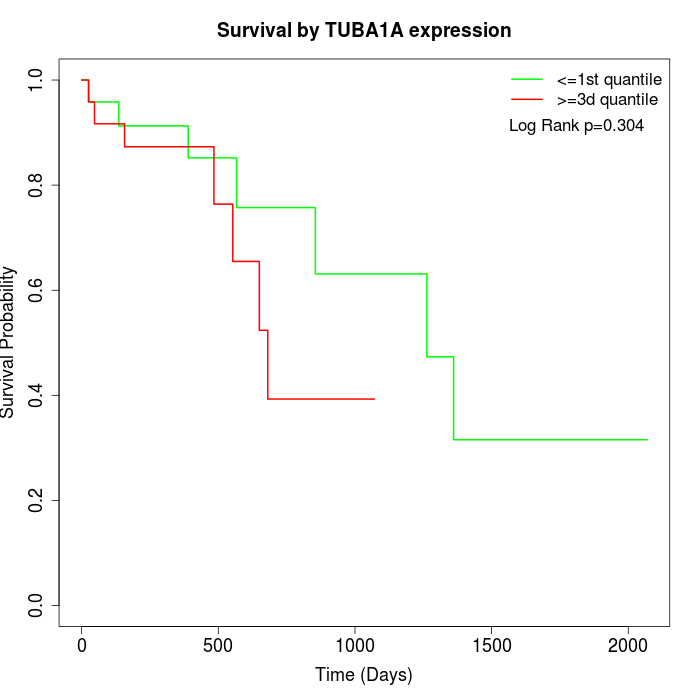
Survival by TUBA1A expression:
Note: Click image to view full size file.
Copy number change of TUBA1A:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TUBA1A | 7846 | 6 | 1 | 23 | |
| GSE20123 | TUBA1A | 7846 | 6 | 1 | 23 | |
| GSE43470 | TUBA1A | 7846 | 4 | 1 | 38 | |
| GSE46452 | TUBA1A | 7846 | 8 | 1 | 50 | |
| GSE47630 | TUBA1A | 7846 | 11 | 2 | 27 | |
| GSE54993 | TUBA1A | 7846 | 0 | 5 | 65 | |
| GSE54994 | TUBA1A | 7846 | 4 | 1 | 48 | |
| GSE60625 | TUBA1A | 7846 | 0 | 0 | 11 | |
| GSE74703 | TUBA1A | 7846 | 4 | 1 | 31 | |
| GSE74704 | TUBA1A | 7846 | 4 | 1 | 15 | |
| TCGA | TUBA1A | 7846 | 16 | 12 | 68 |
Total number of gains: 63; Total number of losses: 26; Total Number of normals: 399.
Somatic mutations of TUBA1A:
Generating mutation plots.
Highly correlated genes for TUBA1A:
Showing top 20/1060 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TUBA1A | C3orf70 | 0.778546 | 3 | 0 | 3 |
| TUBA1A | FERMT2 | 0.768317 | 11 | 0 | 11 |
| TUBA1A | TMEM47 | 0.768134 | 9 | 0 | 9 |
| TUBA1A | CCDC80 | 0.765193 | 4 | 0 | 3 |
| TUBA1A | MGP | 0.751068 | 9 | 0 | 9 |
| TUBA1A | SYNE1 | 0.747362 | 6 | 0 | 6 |
| TUBA1A | TEK | 0.744885 | 3 | 0 | 3 |
| TUBA1A | MYL9 | 0.739401 | 7 | 0 | 7 |
| TUBA1A | SOBP | 0.729979 | 9 | 0 | 9 |
| TUBA1A | MIR100HG | 0.724476 | 4 | 0 | 4 |
| TUBA1A | COMMD6 | 0.723976 | 3 | 0 | 3 |
| TUBA1A | DCHS1 | 0.722216 | 10 | 0 | 10 |
| TUBA1A | FNIP2 | 0.717243 | 4 | 0 | 3 |
| TUBA1A | INMT | 0.715593 | 4 | 0 | 4 |
| TUBA1A | STOM | 0.711564 | 5 | 0 | 5 |
| TUBA1A | MYOCD | 0.709693 | 4 | 0 | 4 |
| TUBA1A | MRVI1 | 0.707599 | 5 | 0 | 4 |
| TUBA1A | EDNRB | 0.705836 | 7 | 0 | 6 |
| TUBA1A | MGARP | 0.705181 | 3 | 0 | 3 |
| TUBA1A | NKAPL | 0.702033 | 3 | 0 | 3 |
For details and further investigation, click here