| Full name: tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon | Alias Symbol: FLJ45465 | ||
| Type: protein-coding gene | Cytoband: 17p13.3 | ||
| Entrez ID: 7531 | HGNC ID: HGNC:12851 | Ensembl Gene: ENSG00000108953 | OMIM ID: 605066 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
YWHAE involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04110 | Cell cycle | |
| hsa04114 | Oocyte meiosis | |
| hsa04151 | PI3K-Akt signaling pathway | |
| hsa04390 | Hippo signaling pathway | |
| hsa04722 | Neurotrophin signaling pathway |
Expression of YWHAE:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | YWHAE | 7531 | 210996_s_at | 0.3256 | 0.3832 | |
| GSE20347 | YWHAE | 7531 | 210996_s_at | 0.1205 | 0.3595 | |
| GSE23400 | YWHAE | 7531 | 210996_s_at | 0.3754 | 0.0000 | |
| GSE26886 | YWHAE | 7531 | 210996_s_at | 0.2973 | 0.1630 | |
| GSE29001 | YWHAE | 7531 | 210996_s_at | 0.0497 | 0.8748 | |
| GSE38129 | YWHAE | 7531 | 210996_s_at | 0.3089 | 0.0918 | |
| GSE45670 | YWHAE | 7531 | 210996_s_at | 0.2383 | 0.0119 | |
| GSE53622 | YWHAE | 7531 | 146310 | 0.1251 | 0.0386 | |
| GSE53624 | YWHAE | 7531 | 15304 | 0.5082 | 0.0000 | |
| GSE63941 | YWHAE | 7531 | 210996_s_at | 1.6202 | 0.0000 | |
| GSE77861 | YWHAE | 7531 | 210996_s_at | 0.0493 | 0.8840 | |
| GSE97050 | YWHAE | 7531 | A_24_P33444 | 0.2246 | 0.3557 | |
| SRP007169 | YWHAE | 7531 | RNAseq | -0.2181 | 0.4398 | |
| SRP008496 | YWHAE | 7531 | RNAseq | -0.1689 | 0.3799 | |
| SRP064894 | YWHAE | 7531 | RNAseq | 0.1261 | 0.4698 | |
| SRP133303 | YWHAE | 7531 | RNAseq | -0.0168 | 0.9130 | |
| SRP159526 | YWHAE | 7531 | RNAseq | 0.2641 | 0.1034 | |
| SRP193095 | YWHAE | 7531 | RNAseq | -0.0836 | 0.5487 | |
| SRP219564 | YWHAE | 7531 | RNAseq | 0.3587 | 0.3109 | |
| TCGA | YWHAE | 7531 | RNAseq | 0.1615 | 0.0000 |
Upregulated datasets: 1; Downregulated datasets: 0.
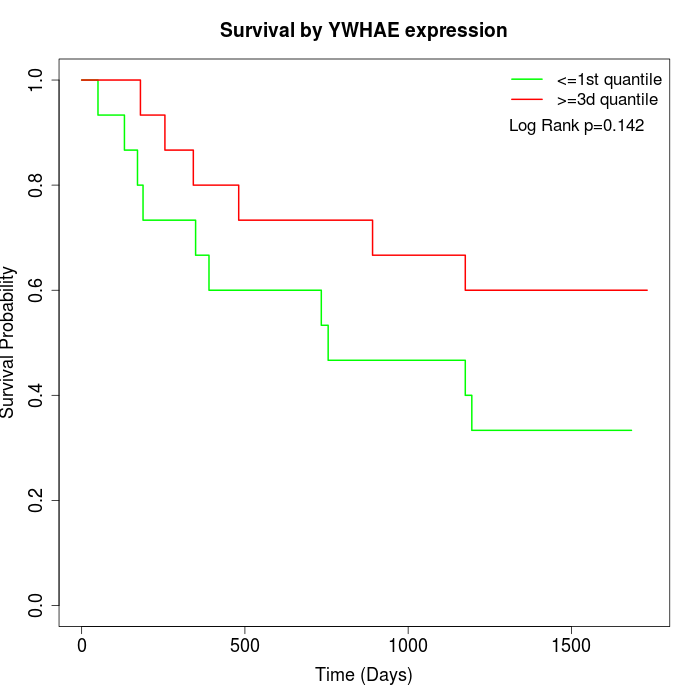
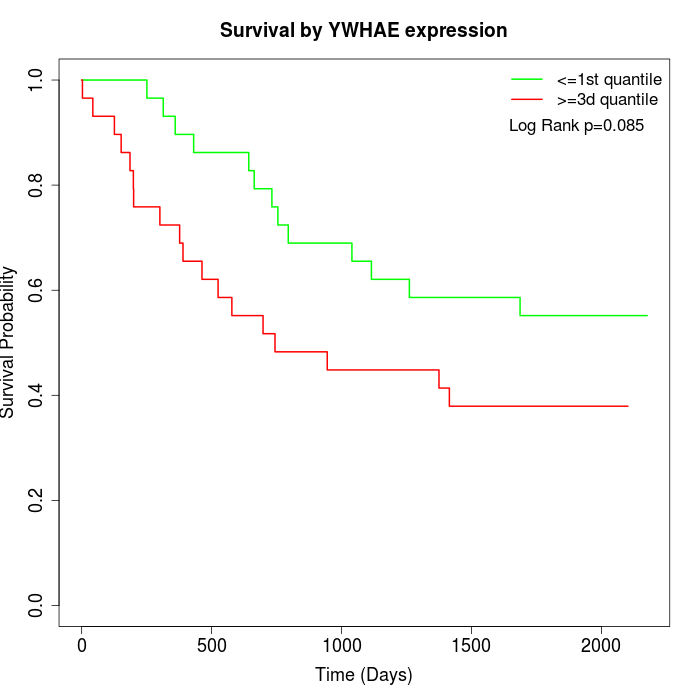
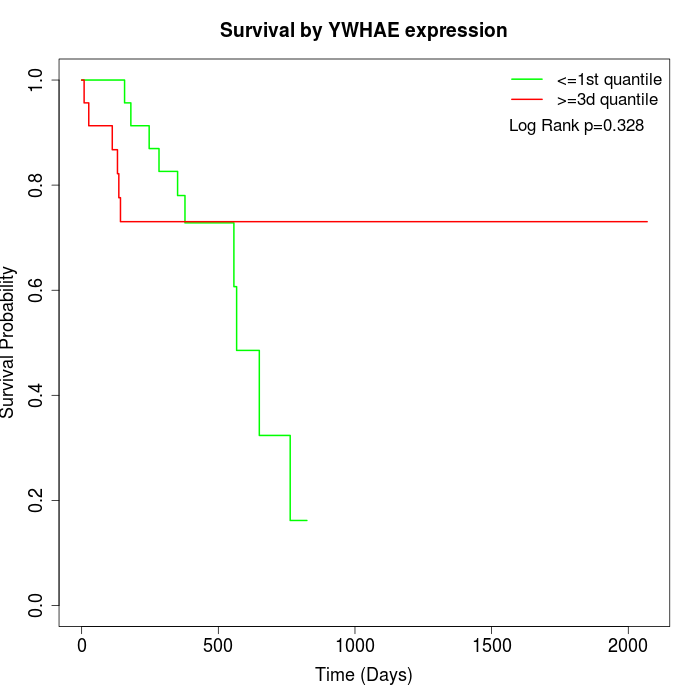
Survival by YWHAE expression:
Note: Click image to view full size file.
Copy number change of YWHAE:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | YWHAE | 7531 | 5 | 2 | 23 | |
| GSE20123 | YWHAE | 7531 | 5 | 3 | 22 | |
| GSE43470 | YWHAE | 7531 | 3 | 6 | 34 | |
| GSE46452 | YWHAE | 7531 | 34 | 1 | 24 | |
| GSE47630 | YWHAE | 7531 | 8 | 1 | 31 | |
| GSE54993 | YWHAE | 7531 | 9 | 3 | 58 | |
| GSE54994 | YWHAE | 7531 | 7 | 9 | 37 | |
| GSE60625 | YWHAE | 7531 | 4 | 0 | 7 | |
| GSE74703 | YWHAE | 7531 | 2 | 3 | 31 | |
| GSE74704 | YWHAE | 7531 | 3 | 1 | 16 | |
| TCGA | YWHAE | 7531 | 19 | 21 | 56 |
Total number of gains: 99; Total number of losses: 50; Total Number of normals: 339.
Somatic mutations of YWHAE:
Generating mutation plots.
Highly correlated genes for YWHAE:
Showing top 20/741 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| YWHAE | CXCL16 | 0.808457 | 3 | 0 | 3 |
| YWHAE | CMTM4 | 0.791761 | 3 | 0 | 3 |
| YWHAE | TMEM167A | 0.781981 | 3 | 0 | 3 |
| YWHAE | TYW5 | 0.770268 | 3 | 0 | 3 |
| YWHAE | PLEKHH1 | 0.770223 | 3 | 0 | 3 |
| YWHAE | PPP2R1B | 0.754963 | 3 | 0 | 3 |
| YWHAE | ARHGAP39 | 0.739502 | 3 | 0 | 3 |
| YWHAE | SCD | 0.738271 | 3 | 0 | 3 |
| YWHAE | CBLC | 0.732293 | 4 | 0 | 3 |
| YWHAE | MARVELD2 | 0.730033 | 4 | 0 | 3 |
| YWHAE | NDUFV1 | 0.719119 | 3 | 0 | 3 |
| YWHAE | SYNE2 | 0.710848 | 4 | 0 | 3 |
| YWHAE | ARHGEF5 | 0.706255 | 4 | 0 | 3 |
| YWHAE | CUL7 | 0.700181 | 5 | 0 | 4 |
| YWHAE | STAP2 | 0.696419 | 3 | 0 | 3 |
| YWHAE | LAMP2 | 0.692784 | 3 | 0 | 3 |
| YWHAE | AP1S3 | 0.689785 | 4 | 0 | 3 |
| YWHAE | NFXL1 | 0.686499 | 3 | 0 | 3 |
| YWHAE | EZH2 | 0.686262 | 5 | 0 | 4 |
| YWHAE | MCMBP | 0.685736 | 4 | 0 | 3 |
For details and further investigation, click here