| Full name: zinc finger CCCH-type containing, antiviral 1 | Alias Symbol: ZAP|FLB6421|FLJ13288|MGC48898|ZC3HDC2|ZC3H2|PARP13 | ||
| Type: protein-coding gene | Cytoband: 7q34 | ||
| Entrez ID: 56829 | HGNC ID: HGNC:23721 | Ensembl Gene: ENSG00000105939 | OMIM ID: 607312 |
| Drug and gene relationship at DGIdb | |||
Expression of ZC3HAV1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ZC3HAV1 | 56829 | 213051_at | 0.2778 | 0.6047 | |
| GSE20347 | ZC3HAV1 | 56829 | 213051_at | 0.2092 | 0.2855 | |
| GSE23400 | ZC3HAV1 | 56829 | 213051_at | 0.2306 | 0.0001 | |
| GSE26886 | ZC3HAV1 | 56829 | 213051_at | 0.0177 | 0.9500 | |
| GSE29001 | ZC3HAV1 | 56829 | 213051_at | 0.2389 | 0.1347 | |
| GSE38129 | ZC3HAV1 | 56829 | 213051_at | 0.0524 | 0.7704 | |
| GSE45670 | ZC3HAV1 | 56829 | 213051_at | 0.3904 | 0.0072 | |
| GSE53622 | ZC3HAV1 | 56829 | 28554 | -0.2618 | 0.0089 | |
| GSE53624 | ZC3HAV1 | 56829 | 61300 | 0.2039 | 0.1012 | |
| GSE63941 | ZC3HAV1 | 56829 | 213051_at | 0.1079 | 0.8367 | |
| GSE77861 | ZC3HAV1 | 56829 | 213051_at | 0.2181 | 0.2928 | |
| GSE97050 | ZC3HAV1 | 56829 | A_33_P3397418 | -0.1339 | 0.5686 | |
| SRP007169 | ZC3HAV1 | 56829 | RNAseq | 0.9836 | 0.0399 | |
| SRP008496 | ZC3HAV1 | 56829 | RNAseq | 0.5326 | 0.0752 | |
| SRP064894 | ZC3HAV1 | 56829 | RNAseq | 0.7202 | 0.0009 | |
| SRP133303 | ZC3HAV1 | 56829 | RNAseq | 0.1912 | 0.2638 | |
| SRP159526 | ZC3HAV1 | 56829 | RNAseq | -0.6322 | 0.0406 | |
| SRP193095 | ZC3HAV1 | 56829 | RNAseq | 0.2047 | 0.0305 | |
| SRP219564 | ZC3HAV1 | 56829 | RNAseq | 0.1415 | 0.7402 | |
| TCGA | ZC3HAV1 | 56829 | RNAseq | -0.0259 | 0.5612 |
Upregulated datasets: 0; Downregulated datasets: 0.
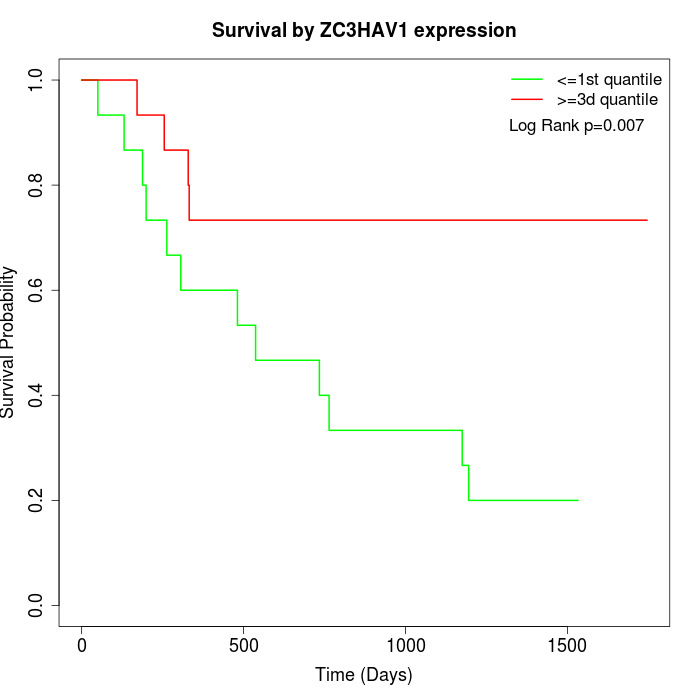
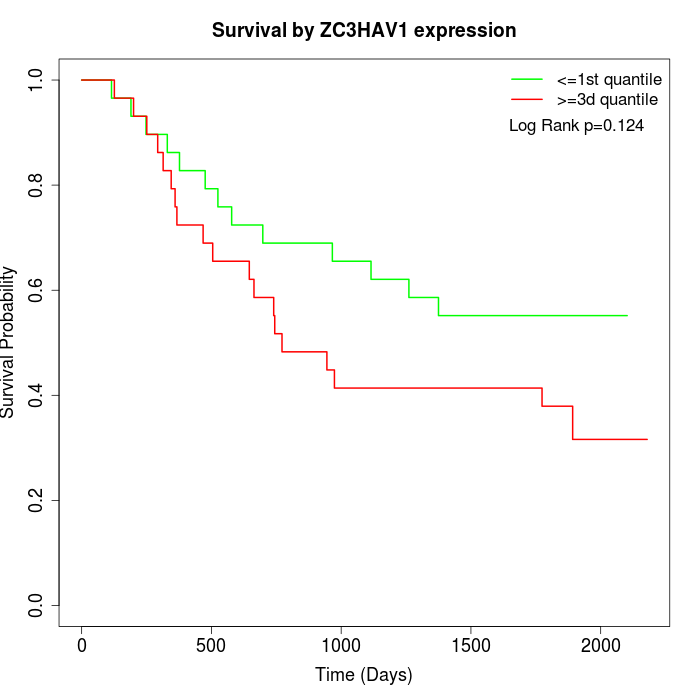
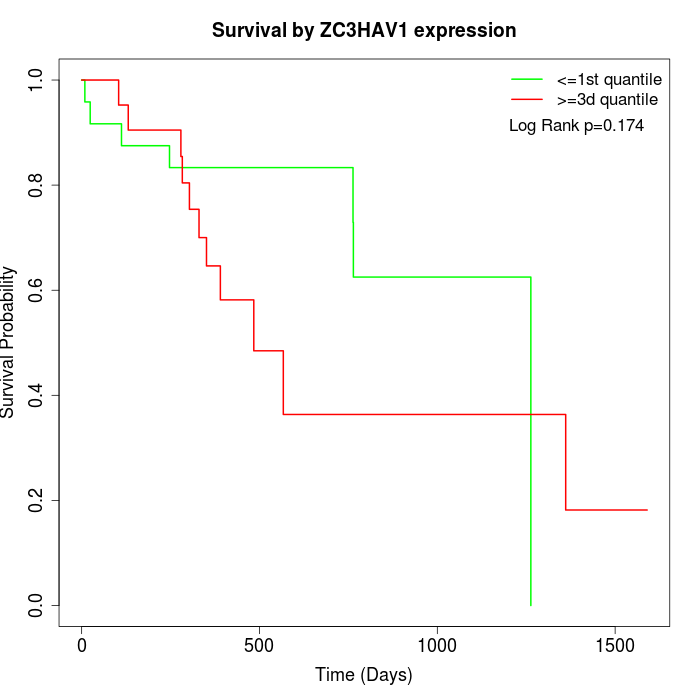
Survival by ZC3HAV1 expression:
Note: Click image to view full size file.
Copy number change of ZC3HAV1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ZC3HAV1 | 56829 | 5 | 2 | 23 | |
| GSE20123 | ZC3HAV1 | 56829 | 5 | 2 | 23 | |
| GSE43470 | ZC3HAV1 | 56829 | 2 | 3 | 38 | |
| GSE46452 | ZC3HAV1 | 56829 | 7 | 2 | 50 | |
| GSE47630 | ZC3HAV1 | 56829 | 7 | 4 | 29 | |
| GSE54993 | ZC3HAV1 | 56829 | 3 | 5 | 62 | |
| GSE54994 | ZC3HAV1 | 56829 | 5 | 8 | 40 | |
| GSE60625 | ZC3HAV1 | 56829 | 0 | 0 | 11 | |
| GSE74703 | ZC3HAV1 | 56829 | 2 | 3 | 31 | |
| GSE74704 | ZC3HAV1 | 56829 | 1 | 2 | 17 | |
| TCGA | ZC3HAV1 | 56829 | 28 | 23 | 45 |
Total number of gains: 65; Total number of losses: 54; Total Number of normals: 369.
Somatic mutations of ZC3HAV1:
Generating mutation plots.
Highly correlated genes for ZC3HAV1:
Showing top 20/242 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ZC3HAV1 | TEX261 | 0.750701 | 3 | 0 | 3 |
| ZC3HAV1 | ERP27 | 0.749984 | 3 | 0 | 3 |
| ZC3HAV1 | ANKRD49 | 0.738804 | 3 | 0 | 3 |
| ZC3HAV1 | ARHGAP35 | 0.730992 | 3 | 0 | 3 |
| ZC3HAV1 | OGT | 0.721768 | 3 | 0 | 3 |
| ZC3HAV1 | C20orf194 | 0.721596 | 3 | 0 | 3 |
| ZC3HAV1 | FAM102B | 0.714249 | 4 | 0 | 4 |
| ZC3HAV1 | JTB | 0.706736 | 4 | 0 | 4 |
| ZC3HAV1 | SLC35A5 | 0.706124 | 3 | 0 | 3 |
| ZC3HAV1 | BTN2A1 | 0.702321 | 3 | 0 | 3 |
| ZC3HAV1 | CASC4 | 0.695323 | 3 | 0 | 3 |
| ZC3HAV1 | SAMD12 | 0.693232 | 3 | 0 | 3 |
| ZC3HAV1 | BRAP | 0.690673 | 3 | 0 | 3 |
| ZC3HAV1 | DAP | 0.689184 | 3 | 0 | 3 |
| ZC3HAV1 | MKLN1 | 0.684159 | 3 | 0 | 3 |
| ZC3HAV1 | TMEM60 | 0.681093 | 3 | 0 | 3 |
| ZC3HAV1 | SNRNP48 | 0.677224 | 3 | 0 | 3 |
| ZC3HAV1 | ARFGEF2 | 0.676097 | 3 | 0 | 3 |
| ZC3HAV1 | CCDC82 | 0.674869 | 3 | 0 | 3 |
| ZC3HAV1 | VPS33A | 0.673773 | 3 | 0 | 3 |
For details and further investigation, click here