| Full name: jumping translocation breakpoint | Alias Symbol: hJT|PAR | ||
| Type: protein-coding gene | Cytoband: 1q21.3 | ||
| Entrez ID: 10899 | HGNC ID: HGNC:6201 | Ensembl Gene: ENSG00000143543 | OMIM ID: 604671 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of JTB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | JTB | 10899 | 200048_s_at | 0.3816 | 0.3422 | |
| GSE20347 | JTB | 10899 | 210434_x_at | 0.4321 | 0.0000 | |
| GSE23400 | JTB | 10899 | 210434_x_at | 0.4468 | 0.0000 | |
| GSE26886 | JTB | 10899 | 200048_s_at | 0.4621 | 0.0063 | |
| GSE29001 | JTB | 10899 | 210434_x_at | 0.3719 | 0.0715 | |
| GSE38129 | JTB | 10899 | 200048_s_at | 0.4786 | 0.0000 | |
| GSE45670 | JTB | 10899 | 200048_s_at | 0.2279 | 0.0893 | |
| GSE53622 | JTB | 10899 | 36247 | 0.1751 | 0.0029 | |
| GSE53624 | JTB | 10899 | 36247 | 0.2817 | 0.0000 | |
| GSE63941 | JTB | 10899 | 210434_x_at | 0.4286 | 0.1688 | |
| GSE77861 | JTB | 10899 | 200048_s_at | 0.5381 | 0.0183 | |
| GSE97050 | JTB | 10899 | A_23_P34983 | 0.3975 | 0.1785 | |
| SRP007169 | JTB | 10899 | RNAseq | -0.4621 | 0.1373 | |
| SRP008496 | JTB | 10899 | RNAseq | 0.0457 | 0.8453 | |
| SRP064894 | JTB | 10899 | RNAseq | 0.0315 | 0.9161 | |
| SRP133303 | JTB | 10899 | RNAseq | 0.2804 | 0.0570 | |
| SRP159526 | JTB | 10899 | RNAseq | 0.4279 | 0.0041 | |
| SRP193095 | JTB | 10899 | RNAseq | -0.1108 | 0.4664 | |
| SRP219564 | JTB | 10899 | RNAseq | 0.4576 | 0.1951 | |
| TCGA | JTB | 10899 | RNAseq | -0.0890 | 0.0652 |
Upregulated datasets: 0; Downregulated datasets: 0.
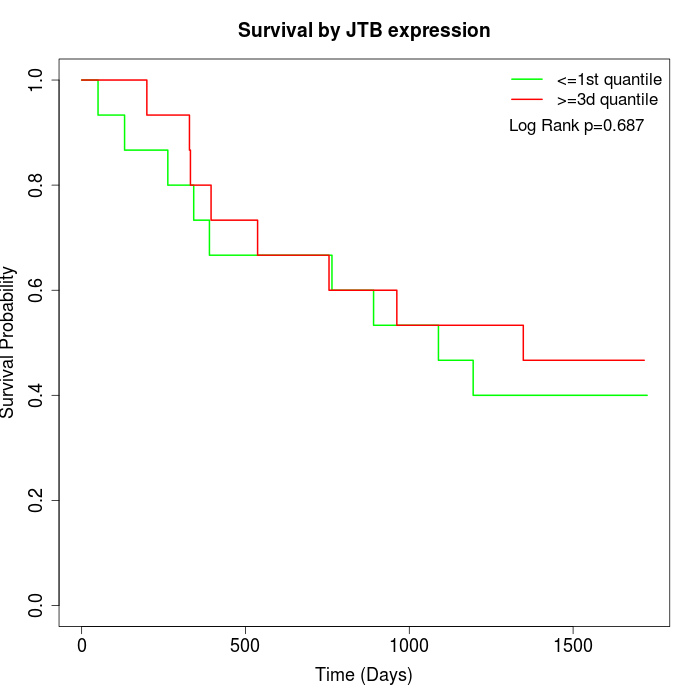
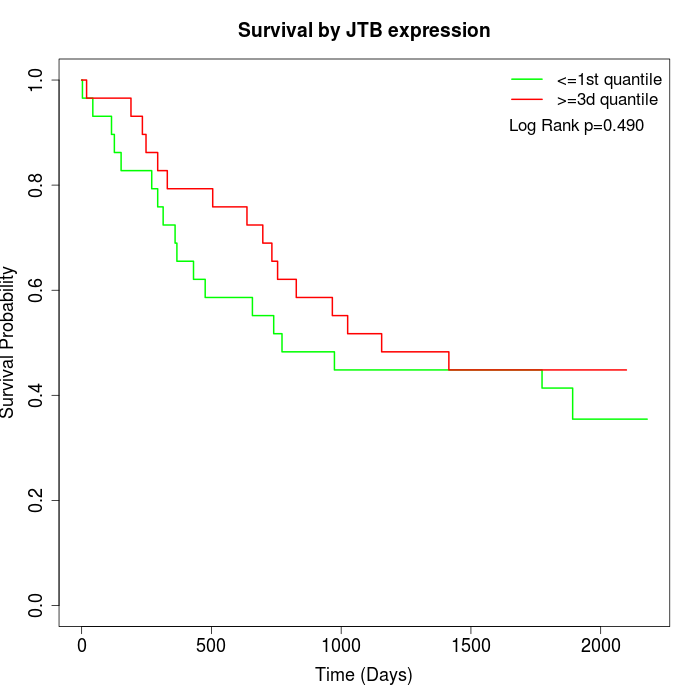
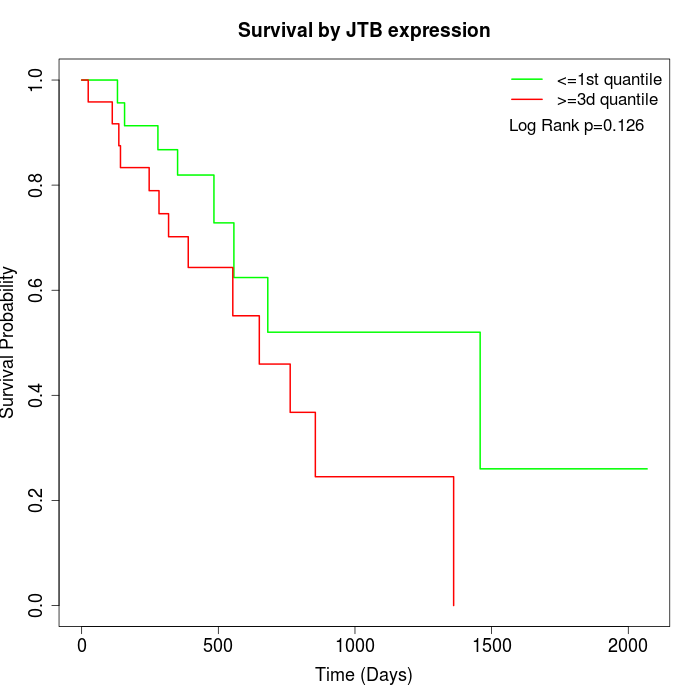
Survival by JTB expression:
Note: Click image to view full size file.
Copy number change of JTB:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | JTB | 10899 | 14 | 0 | 16 | |
| GSE20123 | JTB | 10899 | 14 | 0 | 16 | |
| GSE43470 | JTB | 10899 | 7 | 2 | 34 | |
| GSE46452 | JTB | 10899 | 2 | 1 | 56 | |
| GSE47630 | JTB | 10899 | 14 | 0 | 26 | |
| GSE54993 | JTB | 10899 | 0 | 4 | 66 | |
| GSE54994 | JTB | 10899 | 16 | 0 | 37 | |
| GSE60625 | JTB | 10899 | 0 | 0 | 11 | |
| GSE74703 | JTB | 10899 | 6 | 1 | 29 | |
| GSE74704 | JTB | 10899 | 7 | 0 | 13 | |
| TCGA | JTB | 10899 | 38 | 2 | 56 |
Total number of gains: 118; Total number of losses: 10; Total Number of normals: 360.
Somatic mutations of JTB:
Generating mutation plots.
Highly correlated genes for JTB:
Showing top 20/1633 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| JTB | CYHR1 | 0.821329 | 3 | 0 | 3 |
| JTB | MMACHC | 0.773985 | 3 | 0 | 3 |
| JTB | MADD | 0.771284 | 3 | 0 | 3 |
| JTB | TOR3A | 0.754068 | 10 | 0 | 10 |
| JTB | SESN3 | 0.752461 | 3 | 0 | 3 |
| JTB | DYRK2 | 0.744794 | 5 | 0 | 5 |
| JTB | NXF1 | 0.744749 | 3 | 0 | 3 |
| JTB | KCTD15 | 0.740105 | 4 | 0 | 4 |
| JTB | TTC14 | 0.739823 | 3 | 0 | 3 |
| JTB | SMARCAD1 | 0.739729 | 3 | 0 | 3 |
| JTB | SCO1 | 0.737482 | 3 | 0 | 3 |
| JTB | C12orf73 | 0.735359 | 5 | 0 | 5 |
| JTB | SYNGR3 | 0.733067 | 3 | 0 | 3 |
| JTB | ITGAM | 0.727837 | 3 | 0 | 3 |
| JTB | ARHGAP35 | 0.719766 | 3 | 0 | 3 |
| JTB | NFATC1 | 0.718809 | 3 | 0 | 3 |
| JTB | HYLS1 | 0.717097 | 5 | 0 | 5 |
| JTB | ILF2 | 0.715558 | 12 | 0 | 12 |
| JTB | CERS2 | 0.715227 | 8 | 0 | 8 |
| JTB | TCTN1 | 0.714056 | 3 | 0 | 3 |
For details and further investigation, click here