| Full name: zyg-11 related cell cycle regulator | Alias Symbol: ZYG|Hzyg | ||
| Type: protein-coding gene | Cytoband: 9q34.11 | ||
| Entrez ID: 10444 | HGNC ID: HGNC:30960 | Ensembl Gene: ENSG00000160445 | OMIM ID: 617764 |
| Drug and gene relationship at DGIdb | |||
Expression of ZER1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ZER1 | 10444 | 202452_at | -0.1961 | 0.5843 | |
| GSE20347 | ZER1 | 10444 | 202452_at | -0.2069 | 0.0362 | |
| GSE23400 | ZER1 | 10444 | 202452_at | -0.2005 | 0.0005 | |
| GSE26886 | ZER1 | 10444 | 202452_at | 0.1192 | 0.3803 | |
| GSE29001 | ZER1 | 10444 | 202452_at | -0.0766 | 0.7519 | |
| GSE38129 | ZER1 | 10444 | 202452_at | -0.3516 | 0.0008 | |
| GSE45670 | ZER1 | 10444 | 202452_at | 0.0266 | 0.8030 | |
| GSE53622 | ZER1 | 10444 | 74412 | -0.4794 | 0.0000 | |
| GSE53624 | ZER1 | 10444 | 74412 | -0.2913 | 0.0003 | |
| GSE63941 | ZER1 | 10444 | 202452_at | 0.1764 | 0.5002 | |
| GSE77861 | ZER1 | 10444 | 202452_at | 0.0776 | 0.5922 | |
| GSE97050 | ZER1 | 10444 | A_33_P3256560 | -0.3658 | 0.3440 | |
| SRP007169 | ZER1 | 10444 | RNAseq | -1.0570 | 0.0040 | |
| SRP008496 | ZER1 | 10444 | RNAseq | -0.8365 | 0.0002 | |
| SRP064894 | ZER1 | 10444 | RNAseq | -0.0998 | 0.6823 | |
| SRP133303 | ZER1 | 10444 | RNAseq | -0.3657 | 0.1073 | |
| SRP159526 | ZER1 | 10444 | RNAseq | -0.1102 | 0.6244 | |
| SRP193095 | ZER1 | 10444 | RNAseq | -0.3670 | 0.0029 | |
| SRP219564 | ZER1 | 10444 | RNAseq | -0.6846 | 0.1109 | |
| TCGA | ZER1 | 10444 | RNAseq | -0.1950 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
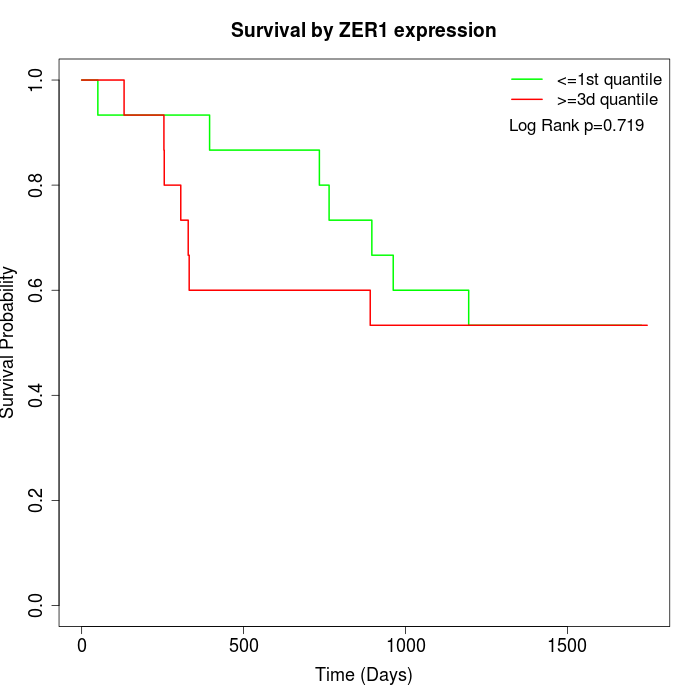
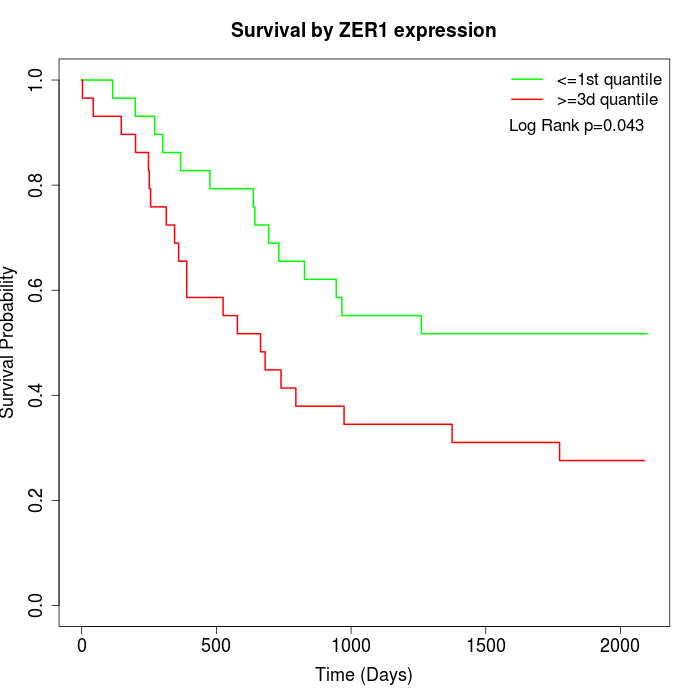
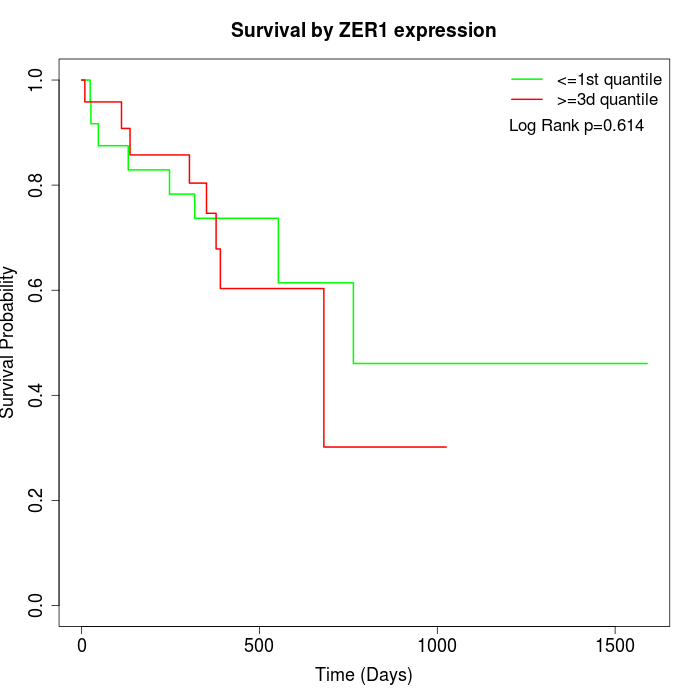
Survival by ZER1 expression:
Note: Click image to view full size file.
Copy number change of ZER1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ZER1 | 10444 | 6 | 9 | 15 | |
| GSE20123 | ZER1 | 10444 | 6 | 9 | 15 | |
| GSE43470 | ZER1 | 10444 | 5 | 8 | 30 | |
| GSE46452 | ZER1 | 10444 | 6 | 13 | 40 | |
| GSE47630 | ZER1 | 10444 | 3 | 16 | 21 | |
| GSE54993 | ZER1 | 10444 | 3 | 3 | 64 | |
| GSE54994 | ZER1 | 10444 | 11 | 8 | 34 | |
| GSE60625 | ZER1 | 10444 | 0 | 0 | 11 | |
| GSE74703 | ZER1 | 10444 | 5 | 6 | 25 | |
| GSE74704 | ZER1 | 10444 | 3 | 7 | 10 | |
| TCGA | ZER1 | 10444 | 27 | 24 | 45 |
Total number of gains: 75; Total number of losses: 103; Total Number of normals: 310.
Somatic mutations of ZER1:
Generating mutation plots.
Highly correlated genes for ZER1:
Showing top 20/709 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ZER1 | RBPMS2 | 0.804663 | 3 | 0 | 3 |
| ZER1 | FBXO30 | 0.802346 | 3 | 0 | 3 |
| ZER1 | C3orf70 | 0.775099 | 3 | 0 | 3 |
| ZER1 | CYP2U1 | 0.767403 | 3 | 0 | 3 |
| ZER1 | SORCS1 | 0.764233 | 3 | 0 | 3 |
| ZER1 | PRIMA1 | 0.745789 | 3 | 0 | 3 |
| ZER1 | FNDC5 | 0.742079 | 3 | 0 | 3 |
| ZER1 | PNCK | 0.737099 | 3 | 0 | 3 |
| ZER1 | STK33 | 0.736449 | 3 | 0 | 3 |
| ZER1 | DDX42 | 0.734765 | 3 | 0 | 3 |
| ZER1 | ADRB3 | 0.732968 | 3 | 0 | 3 |
| ZER1 | RAB9B | 0.728387 | 3 | 0 | 3 |
| ZER1 | KLHL15 | 0.724816 | 3 | 0 | 3 |
| ZER1 | BEND5 | 0.724372 | 4 | 0 | 4 |
| ZER1 | CNN1 | 0.717594 | 4 | 0 | 4 |
| ZER1 | LINC01278 | 0.712936 | 3 | 0 | 3 |
| ZER1 | FRMD3 | 0.711906 | 4 | 0 | 4 |
| ZER1 | RASD2 | 0.709261 | 4 | 0 | 3 |
| ZER1 | DAPK1 | 0.708907 | 3 | 0 | 3 |
| ZER1 | KLHL23 | 0.705216 | 3 | 0 | 3 |
For details and further investigation, click here