| Full name: catenin alpha 2 | Alias Symbol: CAP-R|CT114 | ||
| Type: protein-coding gene | Cytoband: 2p12 | ||
| Entrez ID: 1496 | HGNC ID: HGNC:2510 | Ensembl Gene: ENSG00000066032 | OMIM ID: 114025 |
| Drug and gene relationship at DGIdb | |||
CTNNA2 involved pathways:
Expression of CTNNA2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CTNNA2 | 1496 | 205373_at | -0.2197 | 0.5161 | |
| GSE20347 | CTNNA2 | 1496 | 205373_at | 0.0023 | 0.9713 | |
| GSE23400 | CTNNA2 | 1496 | 205373_at | -0.0047 | 0.8607 | |
| GSE26886 | CTNNA2 | 1496 | 205373_at | 0.0804 | 0.5482 | |
| GSE29001 | CTNNA2 | 1496 | 205373_at | -0.0543 | 0.7094 | |
| GSE38129 | CTNNA2 | 1496 | 205373_at | -0.1533 | 0.0353 | |
| GSE45670 | CTNNA2 | 1496 | 205373_at | -0.1092 | 0.2981 | |
| GSE53622 | CTNNA2 | 1496 | 14079 | 0.2348 | 0.0003 | |
| GSE53624 | CTNNA2 | 1496 | 14079 | 0.2492 | 0.0029 | |
| GSE63941 | CTNNA2 | 1496 | 205373_at | -0.0842 | 0.4608 | |
| GSE77861 | CTNNA2 | 1496 | 205373_at | -0.0684 | 0.4993 | |
| TCGA | CTNNA2 | 1496 | RNAseq | -3.8718 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
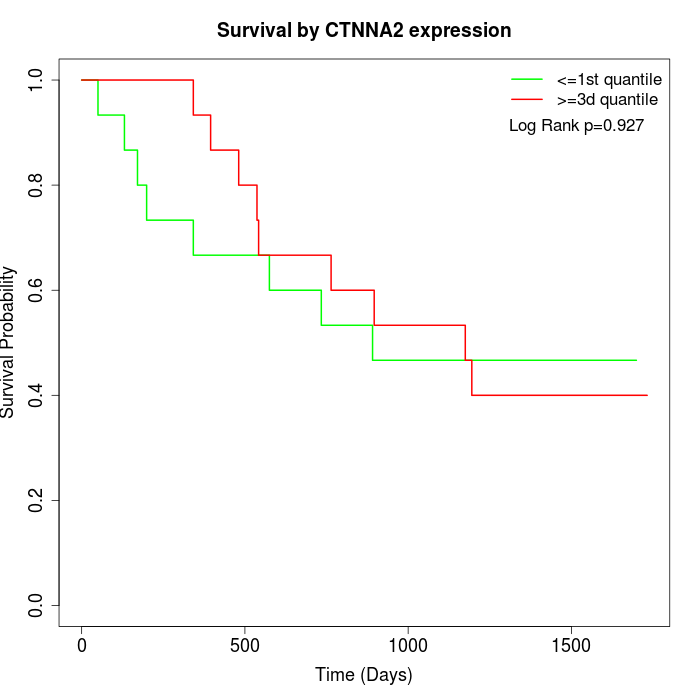
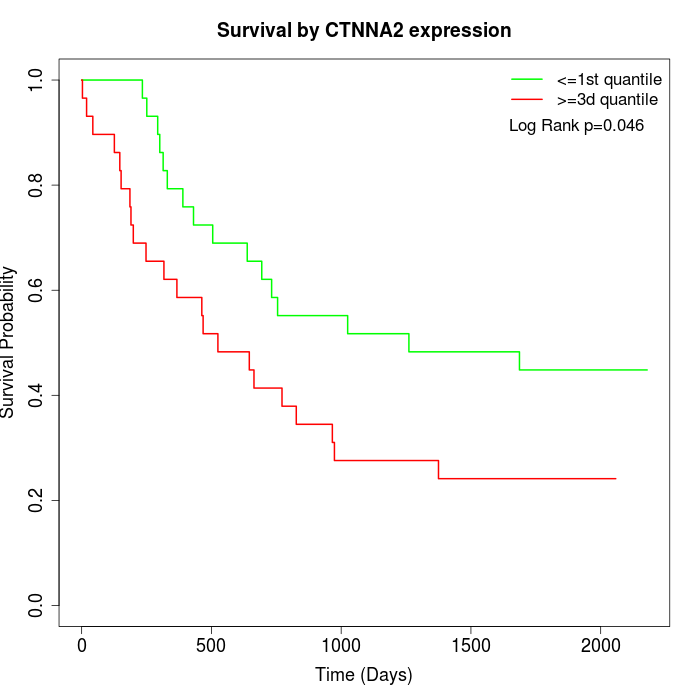
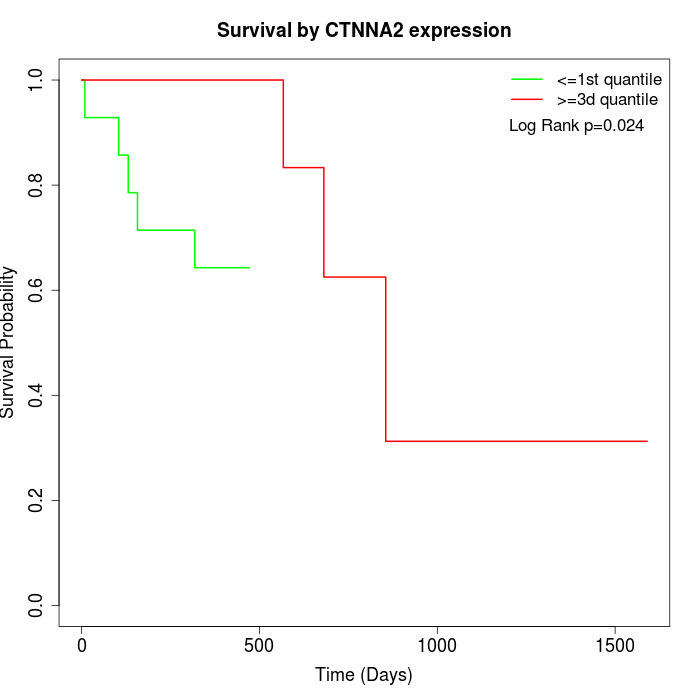
Survival by CTNNA2 expression:
Note: Click image to view full size file.
Copy number change of CTNNA2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CTNNA2 | 1496 | 9 | 2 | 19 | |
| GSE20123 | CTNNA2 | 1496 | 8 | 2 | 20 | |
| GSE43470 | CTNNA2 | 1496 | 5 | 0 | 38 | |
| GSE46452 | CTNNA2 | 1496 | 2 | 3 | 54 | |
| GSE47630 | CTNNA2 | 1496 | 7 | 0 | 33 | |
| GSE54993 | CTNNA2 | 1496 | 0 | 6 | 64 | |
| GSE54994 | CTNNA2 | 1496 | 10 | 0 | 43 | |
| GSE60625 | CTNNA2 | 1496 | 0 | 3 | 8 | |
| GSE74703 | CTNNA2 | 1496 | 5 | 0 | 31 | |
| GSE74704 | CTNNA2 | 1496 | 5 | 1 | 14 | |
| TCGA | CTNNA2 | 1496 | 36 | 6 | 54 |
Total number of gains: 87; Total number of losses: 23; Total Number of normals: 378.
Somatic mutations of CTNNA2:
Generating mutation plots.
Highly correlated genes for CTNNA2:
Showing top 20/207 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CTNNA2 | ZMAT4 | 0.7612 | 3 | 0 | 3 |
| CTNNA2 | MPL | 0.749399 | 3 | 0 | 3 |
| CTNNA2 | MUC8 | 0.745883 | 3 | 0 | 3 |
| CTNNA2 | PLB1 | 0.713162 | 3 | 0 | 3 |
| CTNNA2 | ADAMTS7 | 0.706111 | 3 | 0 | 3 |
| CTNNA2 | PDLIM7 | 0.701721 | 3 | 0 | 3 |
| CTNNA2 | GIP | 0.700856 | 3 | 0 | 3 |
| CTNNA2 | PHEX | 0.696413 | 3 | 0 | 3 |
| CTNNA2 | DNAJB5 | 0.696254 | 3 | 0 | 3 |
| CTNNA2 | LIM2 | 0.684874 | 3 | 0 | 3 |
| CTNNA2 | PSD | 0.673446 | 4 | 0 | 4 |
| CTNNA2 | GNAT3 | 0.671789 | 3 | 0 | 3 |
| CTNNA2 | ADRB3 | 0.66845 | 4 | 0 | 3 |
| CTNNA2 | FOXP3 | 0.664235 | 3 | 0 | 3 |
| CTNNA2 | CDC42EP2 | 0.661759 | 4 | 0 | 3 |
| CTNNA2 | GRIK2 | 0.661488 | 4 | 0 | 4 |
| CTNNA2 | NKX2-8 | 0.652349 | 3 | 0 | 3 |
| CTNNA2 | PRKCG | 0.646145 | 3 | 0 | 3 |
| CTNNA2 | REG3A | 0.645653 | 4 | 0 | 3 |
| CTNNA2 | PNMA3 | 0.645169 | 3 | 0 | 3 |
For details and further investigation, click here