| Full name: zinc finger protein 587 | Alias Symbol: ZF6|FLJ14710|UBF-fl|FLJ20813 | ||
| Type: protein-coding gene | Cytoband: 19q13.43 | ||
| Entrez ID: 84914 | HGNC ID: HGNC:30955 | Ensembl Gene: ENSG00000198466 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of ZNF587:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ZNF587 | 84914 | 219981_x_at | 0.2278 | 0.7852 | |
| GSE20347 | ZNF587 | 84914 | 219981_x_at | -0.0191 | 0.9105 | |
| GSE23400 | ZNF587 | 84914 | 219981_x_at | 0.0064 | 0.9397 | |
| GSE26886 | ZNF587 | 84914 | 219981_x_at | 0.5605 | 0.0070 | |
| GSE29001 | ZNF587 | 84914 | 219981_x_at | 0.1677 | 0.4293 | |
| GSE38129 | ZNF587 | 84914 | 219981_x_at | 0.0261 | 0.8702 | |
| GSE45670 | ZNF587 | 84914 | 219981_x_at | 0.0733 | 0.6066 | |
| GSE53622 | ZNF587 | 84914 | 36278 | 0.1222 | 0.0910 | |
| GSE53624 | ZNF587 | 84914 | 36278 | 0.1312 | 0.1280 | |
| GSE63941 | ZNF587 | 84914 | 219981_x_at | 0.0005 | 0.9992 | |
| GSE77861 | ZNF587 | 84914 | 219981_x_at | 0.2939 | 0.3681 | |
| GSE97050 | ZNF587 | 84914 | A_23_P101281 | 0.0679 | 0.8127 | |
| SRP007169 | ZNF587 | 84914 | RNAseq | 0.1233 | 0.7400 | |
| SRP008496 | ZNF587 | 84914 | RNAseq | 0.1065 | 0.6469 | |
| SRP064894 | ZNF587 | 84914 | RNAseq | -0.0625 | 0.8008 | |
| SRP133303 | ZNF587 | 84914 | RNAseq | 0.1334 | 0.4910 | |
| SRP159526 | ZNF587 | 84914 | RNAseq | 0.1688 | 0.5014 | |
| SRP193095 | ZNF587 | 84914 | RNAseq | 0.0305 | 0.8538 | |
| SRP219564 | ZNF587 | 84914 | RNAseq | 0.1716 | 0.7089 | |
| TCGA | ZNF587 | 84914 | RNAseq | -0.2156 | 0.0008 |
Upregulated datasets: 0; Downregulated datasets: 0.
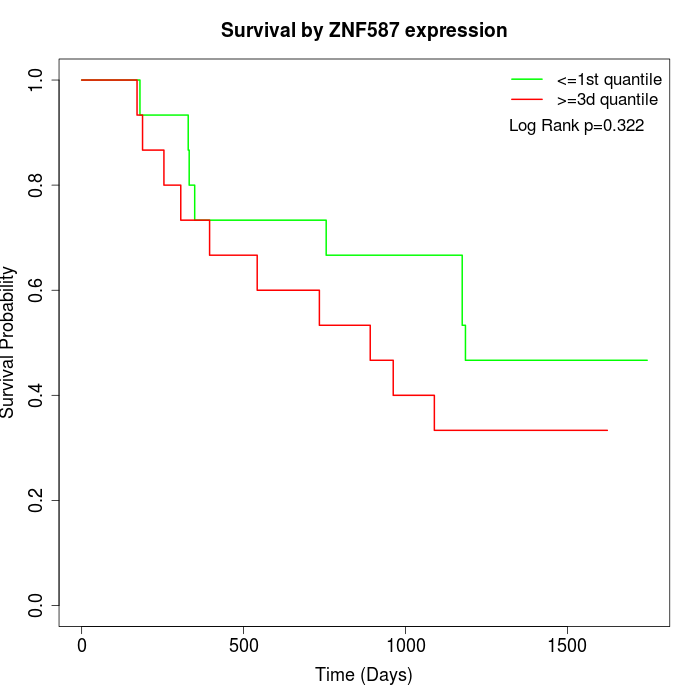
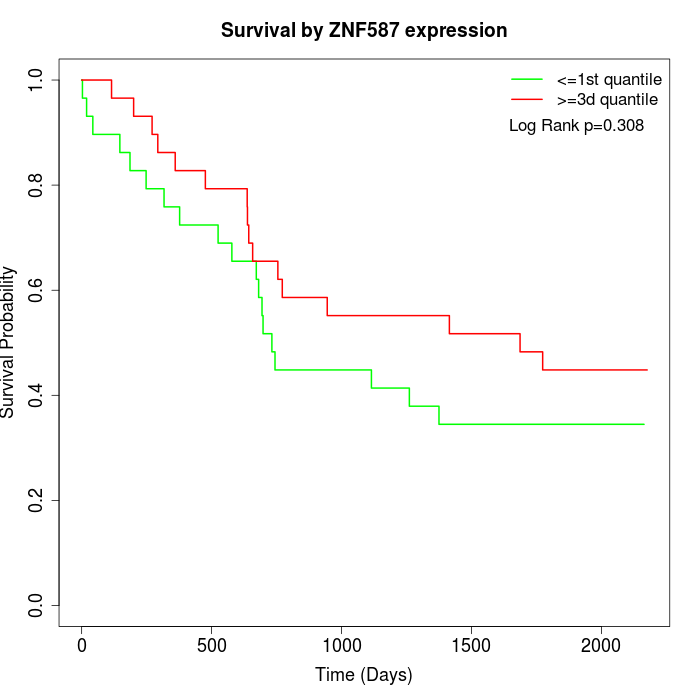
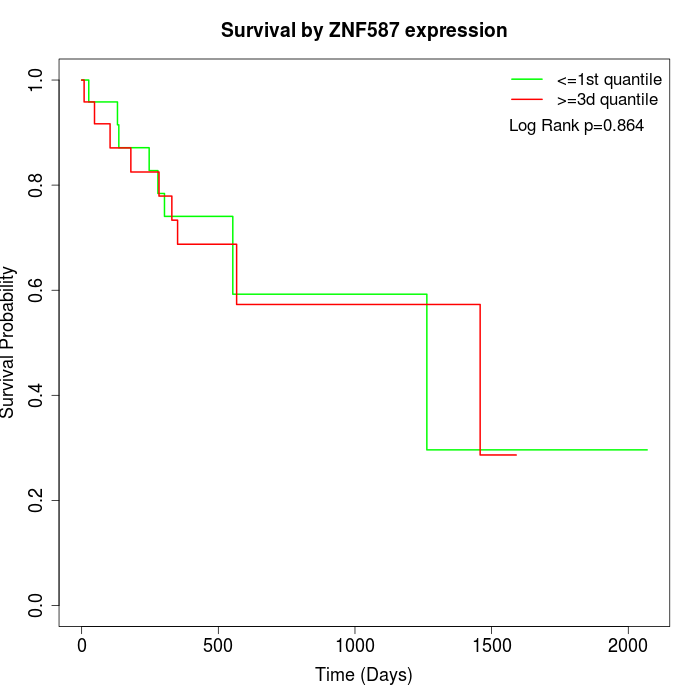
Survival by ZNF587 expression:
Note: Click image to view full size file.
Copy number change of ZNF587:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ZNF587 | 84914 | 2 | 4 | 24 | |
| GSE20123 | ZNF587 | 84914 | 2 | 3 | 25 | |
| GSE43470 | ZNF587 | 84914 | 3 | 9 | 31 | |
| GSE46452 | ZNF587 | 84914 | 45 | 1 | 13 | |
| GSE47630 | ZNF587 | 84914 | 8 | 6 | 26 | |
| GSE54993 | ZNF587 | 84914 | 18 | 4 | 48 | |
| GSE54994 | ZNF587 | 84914 | 5 | 12 | 36 | |
| GSE60625 | ZNF587 | 84914 | 9 | 0 | 2 | |
| GSE74703 | ZNF587 | 84914 | 3 | 6 | 27 | |
| GSE74704 | ZNF587 | 84914 | 2 | 1 | 17 | |
| TCGA | ZNF587 | 84914 | 19 | 15 | 62 |
Total number of gains: 116; Total number of losses: 61; Total Number of normals: 311.
Somatic mutations of ZNF587:
Generating mutation plots.
Highly correlated genes for ZNF587:
Showing top 20/98 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ZNF587 | FAM161B | 0.763962 | 3 | 0 | 3 |
| ZNF587 | C9orf64 | 0.665047 | 4 | 0 | 3 |
| ZNF587 | SLC35E1 | 0.663798 | 7 | 0 | 7 |
| ZNF587 | NLN | 0.656788 | 4 | 0 | 3 |
| ZNF587 | ZNF611 | 0.655941 | 7 | 0 | 7 |
| ZNF587 | HAUS2 | 0.645855 | 7 | 0 | 7 |
| ZNF587 | TRMT10B | 0.643195 | 3 | 0 | 3 |
| ZNF587 | SNW1 | 0.635208 | 3 | 0 | 3 |
| ZNF587 | FBXW12 | 0.633698 | 8 | 0 | 7 |
| ZNF587 | ZC3H7B | 0.631451 | 5 | 0 | 5 |
| ZNF587 | UACA | 0.627905 | 3 | 0 | 3 |
| ZNF587 | FUBP1 | 0.624694 | 3 | 0 | 3 |
| ZNF587 | DIP2A | 0.618045 | 7 | 0 | 7 |
| ZNF587 | UBA2 | 0.617998 | 4 | 0 | 3 |
| ZNF587 | CIC | 0.61541 | 3 | 0 | 3 |
| ZNF587 | RASEF | 0.614389 | 4 | 0 | 3 |
| ZNF587 | GTPBP8 | 0.605231 | 4 | 0 | 3 |
| ZNF587 | VGLL4 | 0.602482 | 4 | 0 | 3 |
| ZNF587 | UBQLN4 | 0.600618 | 5 | 0 | 4 |
| ZNF587 | PUS3 | 0.597912 | 5 | 0 | 4 |
For details and further investigation, click here