| Full name: apoptosis enhancing nuclease | Alias Symbol: FLJ12484|FLJ12562 | ||
| Type: protein-coding gene | Cytoband: 15q26.1 | ||
| Entrez ID: 64782 | HGNC ID: HGNC:25722 | Ensembl Gene: ENSG00000181026 | OMIM ID: 610177 |
| Drug and gene relationship at DGIdb | |||
Expression of AEN:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | AEN | 64782 | 219361_s_at | 0.6542 | 0.0473 | |
| GSE20347 | AEN | 64782 | 219361_s_at | 0.4539 | 0.0103 | |
| GSE23400 | AEN | 64782 | 219361_s_at | 0.2384 | 0.0002 | |
| GSE26886 | AEN | 64782 | 219361_s_at | 0.8656 | 0.0001 | |
| GSE29001 | AEN | 64782 | 219361_s_at | 0.4474 | 0.0930 | |
| GSE38129 | AEN | 64782 | 219361_s_at | 0.3486 | 0.0042 | |
| GSE45670 | AEN | 64782 | 219361_s_at | 0.3865 | 0.0342 | |
| GSE53622 | AEN | 64782 | 3797 | 0.5387 | 0.0000 | |
| GSE53624 | AEN | 64782 | 4534 | 0.6032 | 0.0000 | |
| GSE63941 | AEN | 64782 | 219361_s_at | -0.6915 | 0.1645 | |
| GSE77861 | AEN | 64782 | 219361_s_at | 0.1275 | 0.6057 | |
| GSE97050 | AEN | 64782 | A_33_P3214501 | 0.1637 | 0.5770 | |
| SRP007169 | AEN | 64782 | RNAseq | 0.3522 | 0.3134 | |
| SRP008496 | AEN | 64782 | RNAseq | 0.5396 | 0.0415 | |
| SRP064894 | AEN | 64782 | RNAseq | 0.3713 | 0.0657 | |
| SRP133303 | AEN | 64782 | RNAseq | 0.8214 | 0.0036 | |
| SRP159526 | AEN | 64782 | RNAseq | 0.3424 | 0.3024 | |
| SRP193095 | AEN | 64782 | RNAseq | 0.6917 | 0.0001 | |
| SRP219564 | AEN | 64782 | RNAseq | 0.4508 | 0.4125 | |
| TCGA | AEN | 64782 | RNAseq | -0.0056 | 0.9248 |
Upregulated datasets: 0; Downregulated datasets: 0.
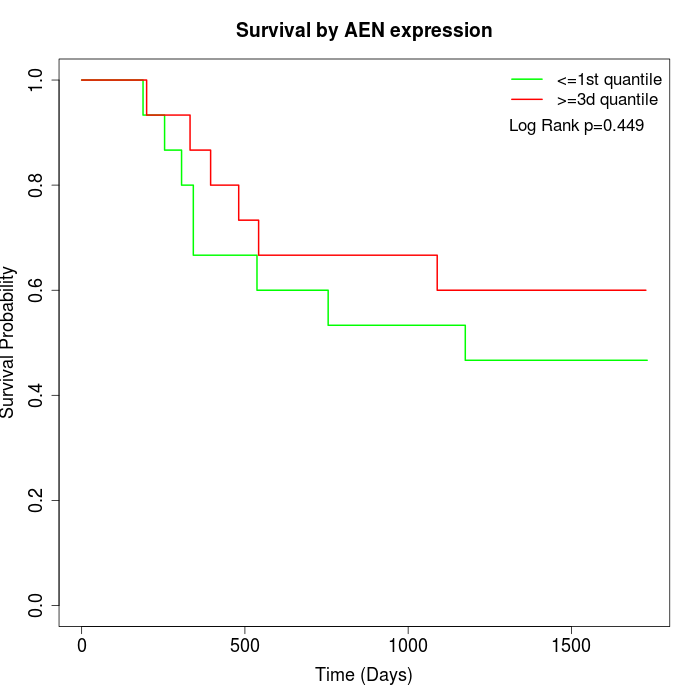
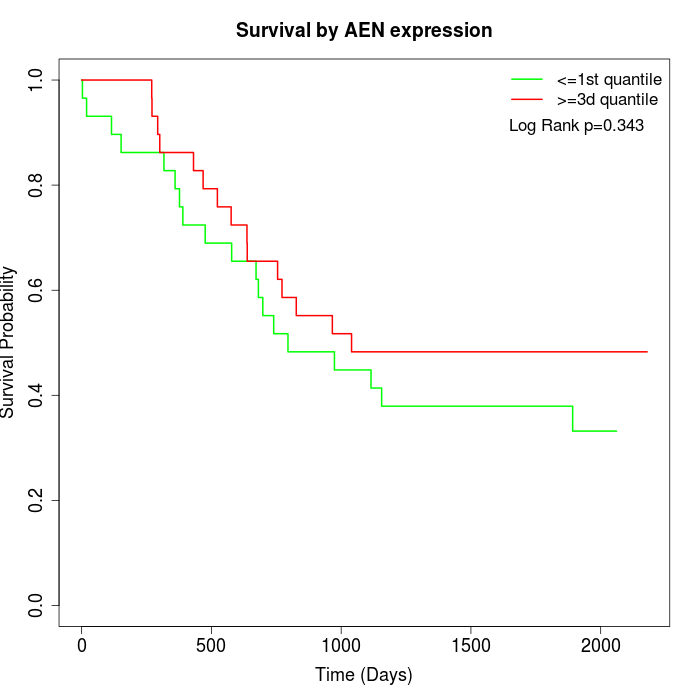
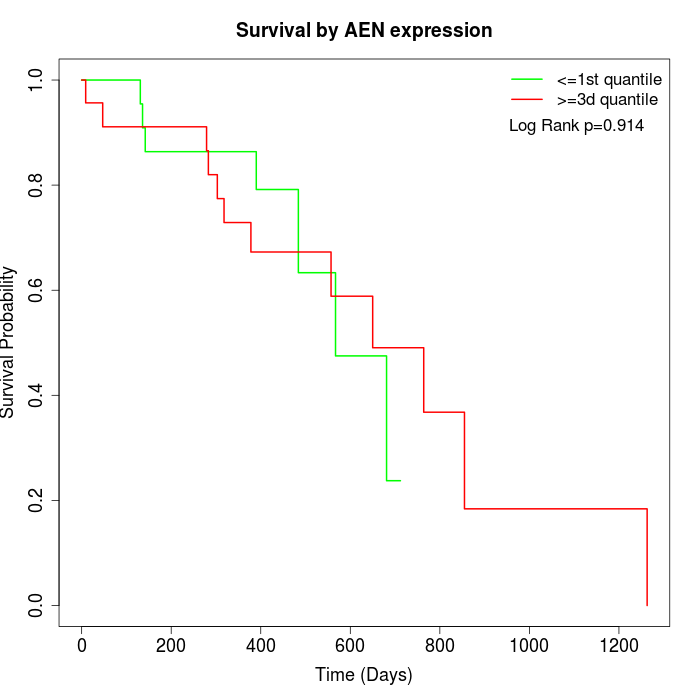
Survival by AEN expression:
Note: Click image to view full size file.
Copy number change of AEN:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | AEN | 64782 | 7 | 3 | 20 | |
| GSE20123 | AEN | 64782 | 7 | 3 | 20 | |
| GSE43470 | AEN | 64782 | 5 | 4 | 34 | |
| GSE46452 | AEN | 64782 | 3 | 7 | 49 | |
| GSE47630 | AEN | 64782 | 8 | 11 | 21 | |
| GSE54993 | AEN | 64782 | 4 | 6 | 60 | |
| GSE54994 | AEN | 64782 | 7 | 6 | 40 | |
| GSE60625 | AEN | 64782 | 4 | 0 | 7 | |
| GSE74703 | AEN | 64782 | 4 | 3 | 29 | |
| GSE74704 | AEN | 64782 | 4 | 2 | 14 | |
| TCGA | AEN | 64782 | 16 | 12 | 68 |
Total number of gains: 69; Total number of losses: 57; Total Number of normals: 362.
Somatic mutations of AEN:
Generating mutation plots.
Highly correlated genes for AEN:
Showing top 20/647 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| AEN | TESK1 | 0.692974 | 3 | 0 | 3 |
| AEN | HAND2 | 0.689378 | 3 | 0 | 3 |
| AEN | TBX1 | 0.679276 | 3 | 0 | 3 |
| AEN | CPT1C | 0.672133 | 4 | 0 | 4 |
| AEN | LIN37 | 0.665452 | 3 | 0 | 3 |
| AEN | RTTN | 0.663326 | 4 | 0 | 3 |
| AEN | DTX3 | 0.662519 | 3 | 0 | 3 |
| AEN | REC8 | 0.660854 | 3 | 0 | 3 |
| AEN | EMD | 0.660647 | 3 | 0 | 3 |
| AEN | WDR5 | 0.658893 | 4 | 0 | 3 |
| AEN | SFXN1 | 0.658047 | 4 | 0 | 4 |
| AEN | TOMM40L | 0.654558 | 3 | 0 | 3 |
| AEN | MZT1 | 0.652664 | 3 | 0 | 3 |
| AEN | ZNF598 | 0.647447 | 4 | 0 | 4 |
| AEN | TEX264 | 0.644966 | 3 | 0 | 3 |
| AEN | MNT | 0.643703 | 4 | 0 | 4 |
| AEN | ROMO1 | 0.643106 | 3 | 0 | 3 |
| AEN | MAP2K5 | 0.642704 | 4 | 0 | 4 |
| AEN | SPRY4-IT1 | 0.642116 | 4 | 0 | 4 |
| AEN | ELFN1 | 0.640584 | 4 | 0 | 4 |
For details and further investigation, click here