| Full name: adenosylhomocysteinase like 1 | Alias Symbol: XPVKONA|IRBIT|PPP1R78 | ||
| Type: protein-coding gene | Cytoband: 1p13.3 | ||
| Entrez ID: 10768 | HGNC ID: HGNC:344 | Ensembl Gene: ENSG00000168710 | OMIM ID: 607826 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of AHCYL1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | AHCYL1 | 10768 | 200850_s_at | -0.4987 | 0.2275 | |
| GSE20347 | AHCYL1 | 10768 | 200850_s_at | -0.4401 | 0.0003 | |
| GSE23400 | AHCYL1 | 10768 | 200850_s_at | -0.0986 | 0.1462 | |
| GSE26886 | AHCYL1 | 10768 | 200850_s_at | -0.5320 | 0.0075 | |
| GSE29001 | AHCYL1 | 10768 | 200849_s_at | -0.4252 | 0.1205 | |
| GSE38129 | AHCYL1 | 10768 | 200850_s_at | -0.3806 | 0.0001 | |
| GSE45670 | AHCYL1 | 10768 | 200850_s_at | -0.2700 | 0.0040 | |
| GSE53622 | AHCYL1 | 10768 | 4362 | -0.3770 | 0.0000 | |
| GSE53624 | AHCYL1 | 10768 | 4362 | -0.2501 | 0.0000 | |
| GSE63941 | AHCYL1 | 10768 | 200850_s_at | 0.0955 | 0.8340 | |
| GSE77861 | AHCYL1 | 10768 | 200850_s_at | -0.1801 | 0.3168 | |
| GSE97050 | AHCYL1 | 10768 | A_24_P11307 | -0.1291 | 0.6006 | |
| SRP007169 | AHCYL1 | 10768 | RNAseq | -0.1950 | 0.5786 | |
| SRP008496 | AHCYL1 | 10768 | RNAseq | -0.6293 | 0.0080 | |
| SRP064894 | AHCYL1 | 10768 | RNAseq | -0.1181 | 0.3614 | |
| SRP133303 | AHCYL1 | 10768 | RNAseq | -0.1719 | 0.1185 | |
| SRP159526 | AHCYL1 | 10768 | RNAseq | -0.4194 | 0.0312 | |
| SRP193095 | AHCYL1 | 10768 | RNAseq | -0.4327 | 0.0000 | |
| SRP219564 | AHCYL1 | 10768 | RNAseq | -0.3066 | 0.3250 | |
| TCGA | AHCYL1 | 10768 | RNAseq | -0.1592 | 0.0001 |
Upregulated datasets: 0; Downregulated datasets: 0.
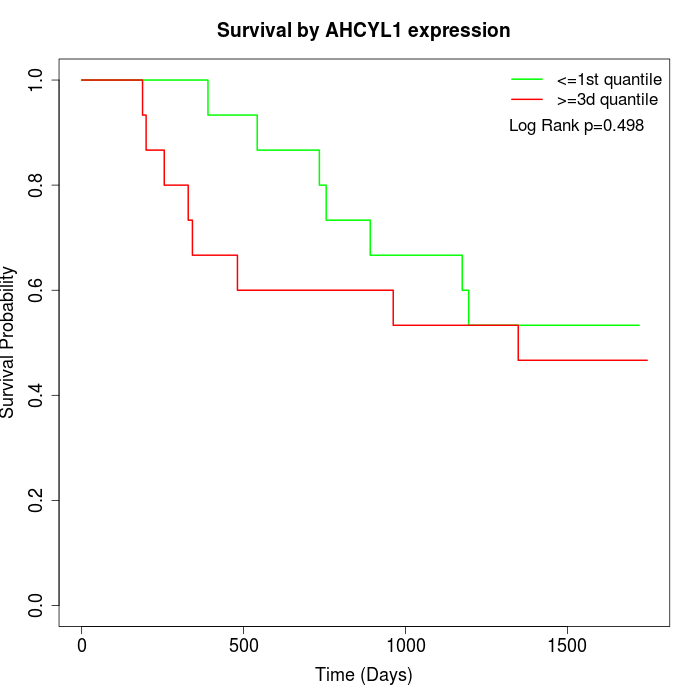
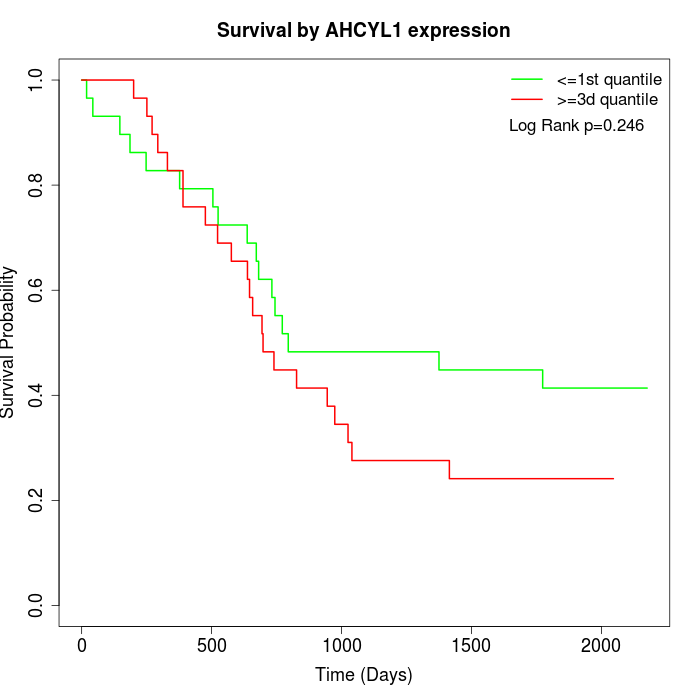
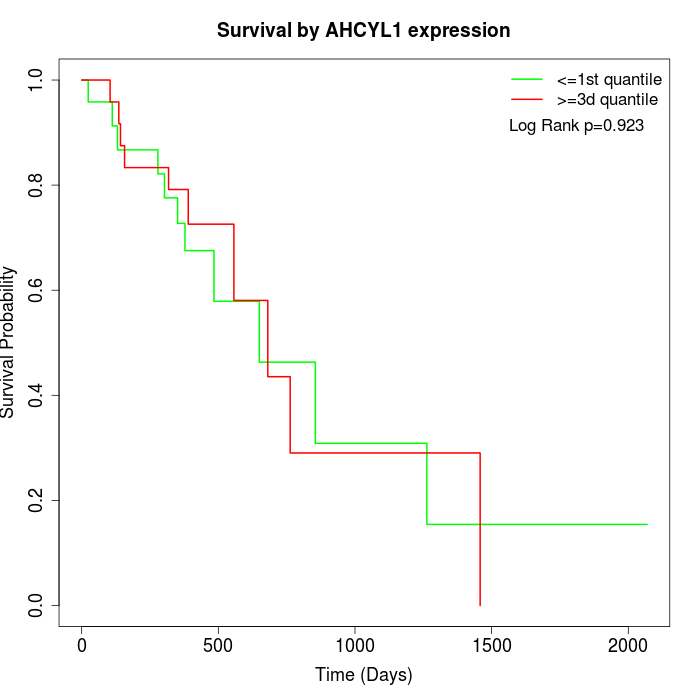
Survival by AHCYL1 expression:
Note: Click image to view full size file.
Copy number change of AHCYL1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | AHCYL1 | 10768 | 0 | 12 | 18 | |
| GSE20123 | AHCYL1 | 10768 | 0 | 11 | 19 | |
| GSE43470 | AHCYL1 | 10768 | 0 | 9 | 34 | |
| GSE46452 | AHCYL1 | 10768 | 2 | 1 | 56 | |
| GSE47630 | AHCYL1 | 10768 | 9 | 5 | 26 | |
| GSE54993 | AHCYL1 | 10768 | 0 | 1 | 69 | |
| GSE54994 | AHCYL1 | 10768 | 7 | 4 | 42 | |
| GSE60625 | AHCYL1 | 10768 | 0 | 0 | 11 | |
| GSE74703 | AHCYL1 | 10768 | 0 | 7 | 29 | |
| GSE74704 | AHCYL1 | 10768 | 0 | 7 | 13 | |
| TCGA | AHCYL1 | 10768 | 9 | 26 | 61 |
Total number of gains: 27; Total number of losses: 83; Total Number of normals: 378.
Somatic mutations of AHCYL1:
Generating mutation plots.
Highly correlated genes for AHCYL1:
Showing top 20/525 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| AHCYL1 | KLHDC9 | 0.703514 | 3 | 0 | 3 |
| AHCYL1 | CNN1 | 0.680655 | 3 | 0 | 3 |
| AHCYL1 | PLCD3 | 0.678854 | 3 | 0 | 3 |
| AHCYL1 | CLCC1 | 0.671288 | 6 | 0 | 6 |
| AHCYL1 | LMO3 | 0.668541 | 3 | 0 | 3 |
| AHCYL1 | CADM2 | 0.668511 | 3 | 0 | 3 |
| AHCYL1 | RHBDD1 | 0.661985 | 3 | 0 | 3 |
| AHCYL1 | KIT | 0.642605 | 4 | 0 | 4 |
| AHCYL1 | ETFA | 0.639343 | 4 | 0 | 3 |
| AHCYL1 | NTN1 | 0.639299 | 5 | 0 | 4 |
| AHCYL1 | SNX15 | 0.637664 | 4 | 0 | 3 |
| AHCYL1 | OGN | 0.635078 | 4 | 0 | 3 |
| AHCYL1 | SNX18 | 0.634299 | 3 | 0 | 3 |
| AHCYL1 | MYOCD | 0.634175 | 4 | 0 | 4 |
| AHCYL1 | MRPL46 | 0.633094 | 3 | 0 | 3 |
| AHCYL1 | TMEM43 | 0.631648 | 6 | 0 | 5 |
| AHCYL1 | SETMAR | 0.630531 | 3 | 0 | 3 |
| AHCYL1 | HMGN1 | 0.627642 | 3 | 0 | 3 |
| AHCYL1 | SLC7A1 | 0.626548 | 4 | 0 | 4 |
| AHCYL1 | CSDE1 | 0.625275 | 8 | 0 | 6 |
For details and further investigation, click here