| Full name: cold shock domain containing E1 | Alias Symbol: D1S155E|UNR | ||
| Type: protein-coding gene | Cytoband: 1p13.2 | ||
| Entrez ID: 7812 | HGNC ID: HGNC:29905 | Ensembl Gene: ENSG00000009307 | OMIM ID: 191510 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of CSDE1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | CSDE1 | 7812 | 219939_s_at | -0.4795 | 0.2706 | |
| GSE20347 | CSDE1 | 7812 | 219939_s_at | -0.6406 | 0.0004 | |
| GSE23400 | CSDE1 | 7812 | 202646_s_at | -0.2732 | 0.0037 | |
| GSE26886 | CSDE1 | 7812 | 222975_s_at | -1.6414 | 0.0000 | |
| GSE29001 | CSDE1 | 7812 | 202646_s_at | -0.0805 | 0.8266 | |
| GSE38129 | CSDE1 | 7812 | 219939_s_at | -0.6950 | 0.0000 | |
| GSE45670 | CSDE1 | 7812 | 219939_s_at | -0.5872 | 0.0000 | |
| GSE53622 | CSDE1 | 7812 | 2349 | -0.8844 | 0.0000 | |
| GSE53624 | CSDE1 | 7812 | 2349 | -0.5284 | 0.0000 | |
| GSE63941 | CSDE1 | 7812 | 219939_s_at | -0.6103 | 0.1868 | |
| GSE77861 | CSDE1 | 7812 | 219939_s_at | -0.6219 | 0.0101 | |
| GSE97050 | CSDE1 | 7812 | A_32_P73821 | -0.5835 | 0.1110 | |
| SRP007169 | CSDE1 | 7812 | RNAseq | -0.0893 | 0.8056 | |
| SRP008496 | CSDE1 | 7812 | RNAseq | -0.1822 | 0.3892 | |
| SRP064894 | CSDE1 | 7812 | RNAseq | -0.3132 | 0.0130 | |
| SRP133303 | CSDE1 | 7812 | RNAseq | -0.1864 | 0.2641 | |
| SRP159526 | CSDE1 | 7812 | RNAseq | -0.6312 | 0.0009 | |
| SRP193095 | CSDE1 | 7812 | RNAseq | -0.5215 | 0.0000 | |
| SRP219564 | CSDE1 | 7812 | RNAseq | -0.8065 | 0.1483 | |
| TCGA | CSDE1 | 7812 | RNAseq | -0.1395 | 0.0003 |
Upregulated datasets: 0; Downregulated datasets: 1.
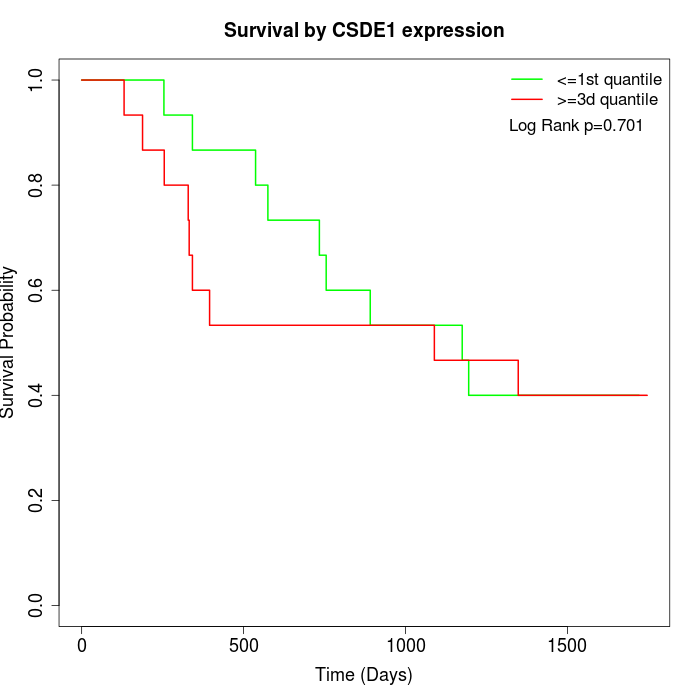
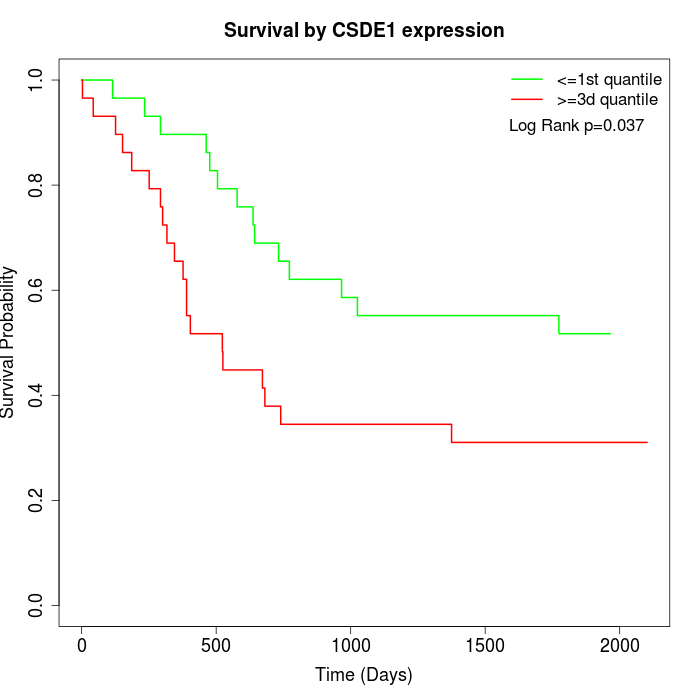
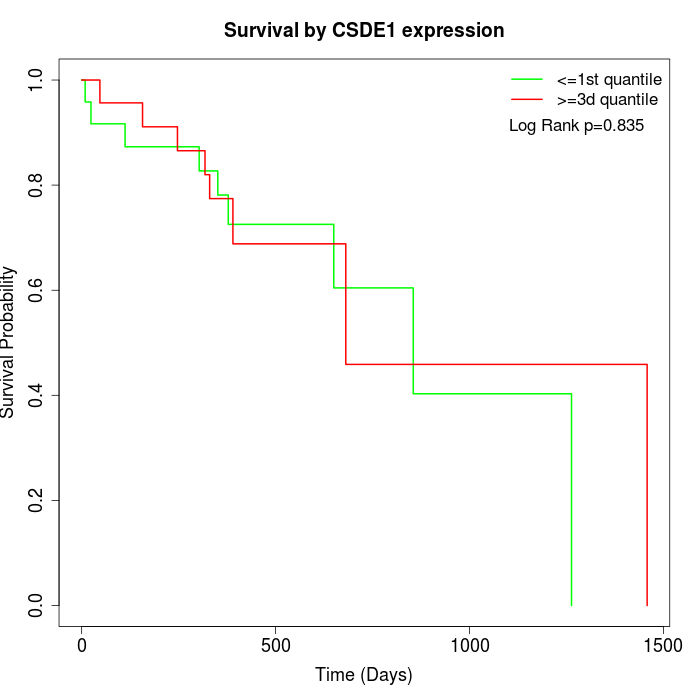
Survival by CSDE1 expression:
Note: Click image to view full size file.
Copy number change of CSDE1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | CSDE1 | 7812 | 0 | 9 | 21 | |
| GSE20123 | CSDE1 | 7812 | 0 | 9 | 21 | |
| GSE43470 | CSDE1 | 7812 | 0 | 8 | 35 | |
| GSE46452 | CSDE1 | 7812 | 2 | 1 | 56 | |
| GSE47630 | CSDE1 | 7812 | 9 | 5 | 26 | |
| GSE54993 | CSDE1 | 7812 | 0 | 1 | 69 | |
| GSE54994 | CSDE1 | 7812 | 7 | 3 | 43 | |
| GSE60625 | CSDE1 | 7812 | 0 | 0 | 11 | |
| GSE74703 | CSDE1 | 7812 | 0 | 7 | 29 | |
| GSE74704 | CSDE1 | 7812 | 0 | 5 | 15 | |
| TCGA | CSDE1 | 7812 | 13 | 30 | 53 |
Total number of gains: 31; Total number of losses: 78; Total Number of normals: 379.
Somatic mutations of CSDE1:
Generating mutation plots.
Highly correlated genes for CSDE1:
Showing top 20/1323 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| CSDE1 | PREX2 | 0.822058 | 3 | 0 | 3 |
| CSDE1 | CYP4V2 | 0.790187 | 3 | 0 | 3 |
| CSDE1 | AKAP6 | 0.787419 | 3 | 0 | 3 |
| CSDE1 | SLMAP | 0.769732 | 7 | 0 | 6 |
| CSDE1 | EIF4E3 | 0.755336 | 7 | 0 | 6 |
| CSDE1 | BLOC1S2 | 0.750513 | 3 | 0 | 3 |
| CSDE1 | TMEM234 | 0.750286 | 3 | 0 | 3 |
| CSDE1 | MRPL39 | 0.748988 | 5 | 0 | 5 |
| CSDE1 | ANAPC16 | 0.747319 | 5 | 0 | 4 |
| CSDE1 | HIPK1 | 0.746769 | 10 | 0 | 9 |
| CSDE1 | CLEC3B | 0.74638 | 3 | 0 | 3 |
| CSDE1 | RPP30 | 0.744246 | 3 | 0 | 3 |
| CSDE1 | MTFR1L | 0.741288 | 5 | 0 | 5 |
| CSDE1 | TMEM218 | 0.740892 | 3 | 0 | 3 |
| CSDE1 | SEC24C | 0.740095 | 3 | 0 | 3 |
| CSDE1 | PDK3 | 0.737495 | 4 | 0 | 3 |
| CSDE1 | OIP5-AS1 | 0.735873 | 3 | 0 | 3 |
| CSDE1 | BDH2 | 0.735526 | 6 | 0 | 6 |
| CSDE1 | MACROD2 | 0.735455 | 3 | 0 | 3 |
| CSDE1 | FAM83D | 0.733407 | 4 | 0 | 4 |
For details and further investigation, click here