| Full name: SET domain and mariner transposase fusion gene | Alias Symbol: metnase | ||
| Type: protein-coding gene | Cytoband: 3p26.1 | ||
| Entrez ID: 6419 | HGNC ID: HGNC:10762 | Ensembl Gene: ENSG00000170364 | OMIM ID: 609834 |
| Drug and gene relationship at DGIdb | |||
Expression of SETMAR:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | SETMAR | 6419 | 206554_x_at | -0.7869 | 0.2656 | |
| GSE20347 | SETMAR | 6419 | 206554_x_at | -0.4606 | 0.0187 | |
| GSE23400 | SETMAR | 6419 | 206554_x_at | -0.1717 | 0.0000 | |
| GSE26886 | SETMAR | 6419 | 206554_x_at | -0.0893 | 0.8280 | |
| GSE29001 | SETMAR | 6419 | 206554_x_at | -0.5601 | 0.0777 | |
| GSE38129 | SETMAR | 6419 | 206554_x_at | -0.6810 | 0.0000 | |
| GSE45670 | SETMAR | 6419 | 206554_x_at | -0.9711 | 0.0002 | |
| GSE53622 | SETMAR | 6419 | 28590 | -0.0526 | 0.5681 | |
| GSE53624 | SETMAR | 6419 | 28590 | -0.1793 | 0.0005 | |
| GSE63941 | SETMAR | 6419 | 206554_x_at | -1.1138 | 0.1654 | |
| GSE77861 | SETMAR | 6419 | 206554_x_at | -0.6877 | 0.0039 | |
| GSE97050 | SETMAR | 6419 | A_23_P80643 | -0.1203 | 0.5580 | |
| SRP007169 | SETMAR | 6419 | RNAseq | -1.1743 | 0.0011 | |
| SRP008496 | SETMAR | 6419 | RNAseq | -0.4471 | 0.1515 | |
| SRP064894 | SETMAR | 6419 | RNAseq | -0.5088 | 0.0256 | |
| SRP133303 | SETMAR | 6419 | RNAseq | -0.2893 | 0.0004 | |
| SRP159526 | SETMAR | 6419 | RNAseq | -0.4274 | 0.1894 | |
| SRP193095 | SETMAR | 6419 | RNAseq | -0.2407 | 0.0353 | |
| SRP219564 | SETMAR | 6419 | RNAseq | -1.0934 | 0.0059 | |
| TCGA | SETMAR | 6419 | RNAseq | -0.2617 | 0.0074 |
Upregulated datasets: 0; Downregulated datasets: 2.
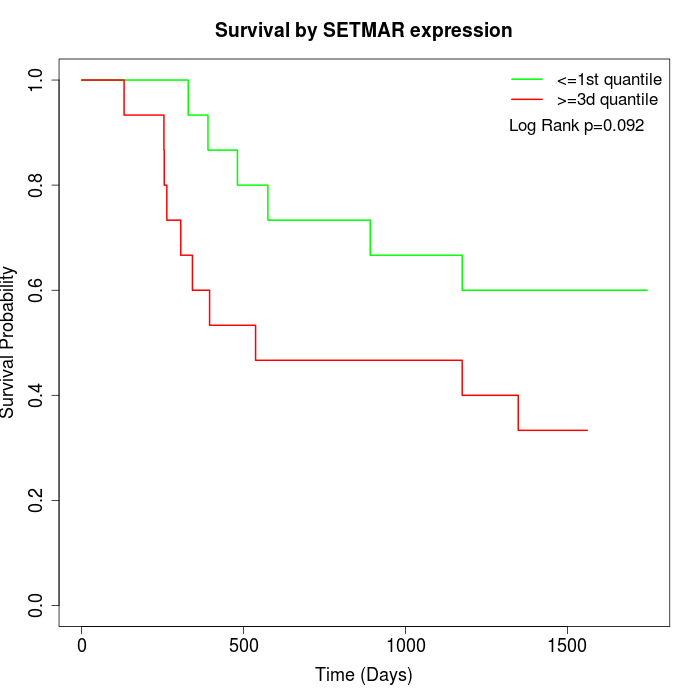
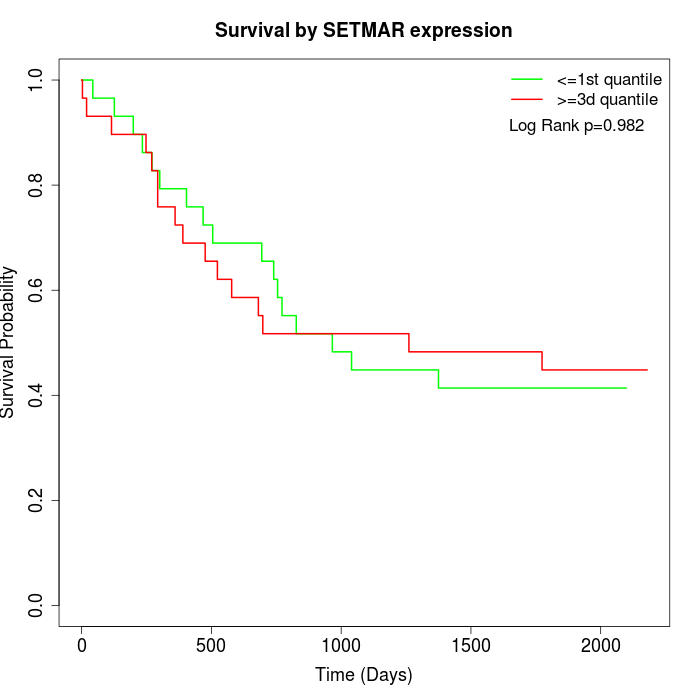
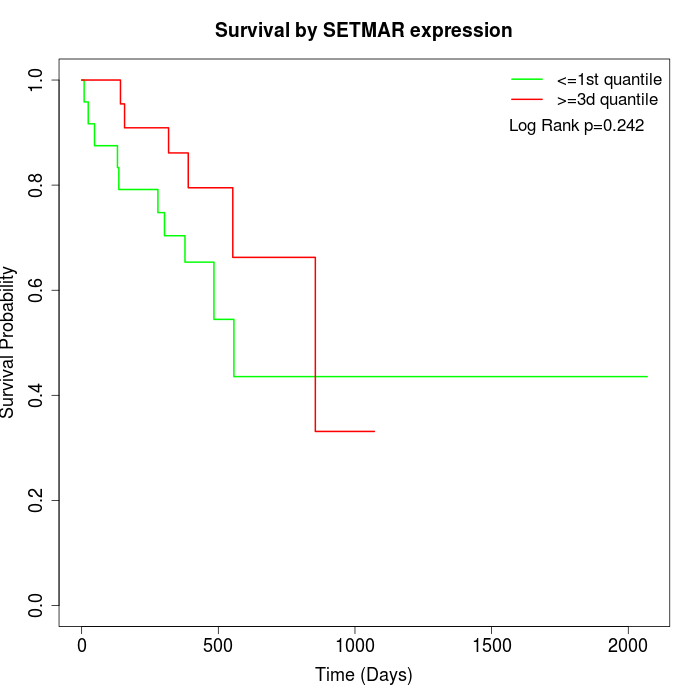
Survival by SETMAR expression:
Note: Click image to view full size file.
Copy number change of SETMAR:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | SETMAR | 6419 | 1 | 19 | 10 | |
| GSE20123 | SETMAR | 6419 | 1 | 19 | 10 | |
| GSE43470 | SETMAR | 6419 | 0 | 21 | 22 | |
| GSE46452 | SETMAR | 6419 | 2 | 18 | 39 | |
| GSE47630 | SETMAR | 6419 | 1 | 24 | 15 | |
| GSE54993 | SETMAR | 6419 | 6 | 1 | 63 | |
| GSE54994 | SETMAR | 6419 | 1 | 32 | 20 | |
| GSE60625 | SETMAR | 6419 | 5 | 0 | 6 | |
| GSE74703 | SETMAR | 6419 | 0 | 18 | 18 | |
| GSE74704 | SETMAR | 6419 | 1 | 11 | 8 | |
| TCGA | SETMAR | 6419 | 2 | 66 | 28 |
Total number of gains: 20; Total number of losses: 229; Total Number of normals: 239.
Somatic mutations of SETMAR:
Generating mutation plots.
Highly correlated genes for SETMAR:
Showing top 20/863 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| SETMAR | SVIP | 0.763669 | 5 | 0 | 5 |
| SETMAR | ZNF223 | 0.763277 | 3 | 0 | 3 |
| SETMAR | ZNF823 | 0.761411 | 3 | 0 | 3 |
| SETMAR | FAM200B | 0.754744 | 3 | 0 | 3 |
| SETMAR | SLC46A2 | 0.747152 | 3 | 0 | 3 |
| SETMAR | ECH1 | 0.742434 | 3 | 0 | 3 |
| SETMAR | NKAPL | 0.742019 | 3 | 0 | 3 |
| SETMAR | MPPED2 | 0.73998 | 3 | 0 | 3 |
| SETMAR | ZNF436 | 0.739729 | 3 | 0 | 3 |
| SETMAR | NIPSNAP3A | 0.739533 | 4 | 0 | 4 |
| SETMAR | GNPDA2 | 0.7393 | 3 | 0 | 3 |
| SETMAR | TAF7 | 0.735935 | 3 | 0 | 3 |
| SETMAR | DOCK8 | 0.735884 | 3 | 0 | 3 |
| SETMAR | POGK | 0.735303 | 3 | 0 | 3 |
| SETMAR | CETN3 | 0.732436 | 4 | 0 | 3 |
| SETMAR | RPL32 | 0.731645 | 8 | 0 | 8 |
| SETMAR | EIF4E3 | 0.731347 | 4 | 0 | 4 |
| SETMAR | SLC30A4 | 0.729173 | 3 | 0 | 3 |
| SETMAR | RNF135 | 0.729024 | 3 | 0 | 3 |
| SETMAR | NFIA | 0.72613 | 5 | 0 | 4 |
For details and further investigation, click here