| Full name: ALG5 dolichyl-phosphate beta-glucosyltransferase | Alias Symbol: bA421P11.2 | ||
| Type: protein-coding gene | Cytoband: 13q13.3 | ||
| Entrez ID: 29880 | HGNC ID: HGNC:20266 | Ensembl Gene: ENSG00000120697 | OMIM ID: 604565 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of ALG5:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ALG5 | 29880 | 218203_at | 0.0920 | 0.8437 | |
| GSE20347 | ALG5 | 29880 | 218203_at | 0.0371 | 0.7862 | |
| GSE23400 | ALG5 | 29880 | 218203_at | 0.3313 | 0.0003 | |
| GSE26886 | ALG5 | 29880 | 218203_at | 0.1887 | 0.5538 | |
| GSE29001 | ALG5 | 29880 | 218203_at | 0.0867 | 0.8528 | |
| GSE38129 | ALG5 | 29880 | 218203_at | 0.1629 | 0.1902 | |
| GSE45670 | ALG5 | 29880 | 218203_at | 0.0923 | 0.4865 | |
| GSE53622 | ALG5 | 29880 | 49345 | -0.0060 | 0.9409 | |
| GSE53624 | ALG5 | 29880 | 49345 | 0.1869 | 0.0070 | |
| GSE63941 | ALG5 | 29880 | 218203_at | -0.3756 | 0.3091 | |
| GSE77861 | ALG5 | 29880 | 218203_at | 0.1285 | 0.6703 | |
| GSE97050 | ALG5 | 29880 | A_23_P151436 | 0.2344 | 0.2873 | |
| SRP007169 | ALG5 | 29880 | RNAseq | -0.5758 | 0.1223 | |
| SRP008496 | ALG5 | 29880 | RNAseq | -0.3897 | 0.1204 | |
| SRP064894 | ALG5 | 29880 | RNAseq | 0.4147 | 0.0533 | |
| SRP133303 | ALG5 | 29880 | RNAseq | 0.1193 | 0.5018 | |
| SRP159526 | ALG5 | 29880 | RNAseq | -0.0597 | 0.8328 | |
| SRP193095 | ALG5 | 29880 | RNAseq | -0.3566 | 0.0146 | |
| SRP219564 | ALG5 | 29880 | RNAseq | 0.0318 | 0.9166 | |
| TCGA | ALG5 | 29880 | RNAseq | -0.2161 | 0.0002 |
Upregulated datasets: 0; Downregulated datasets: 0.
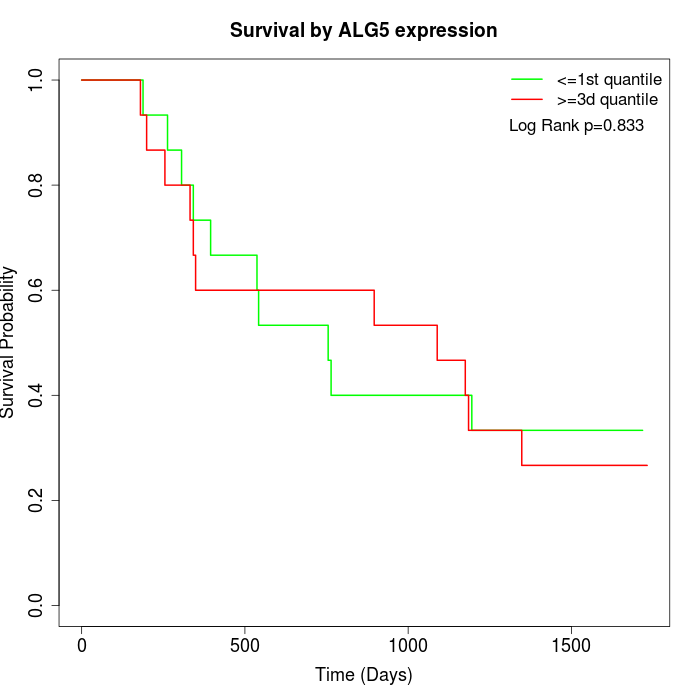
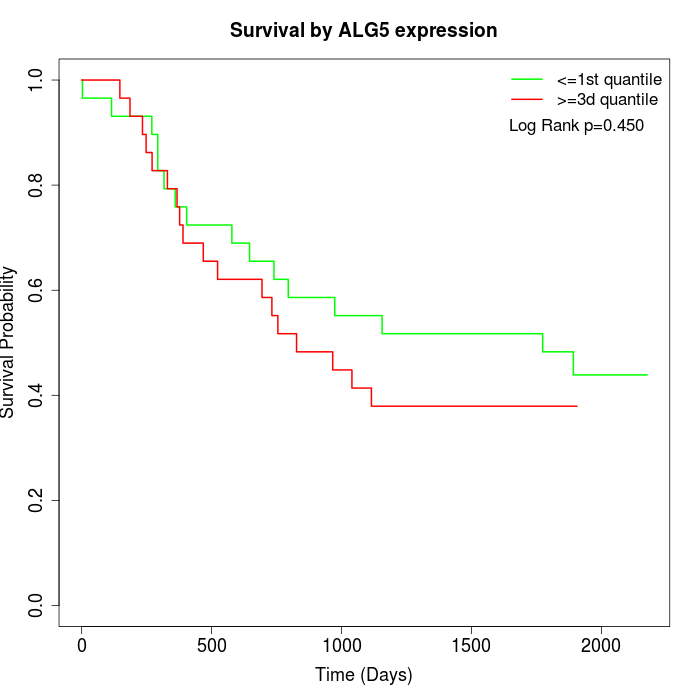
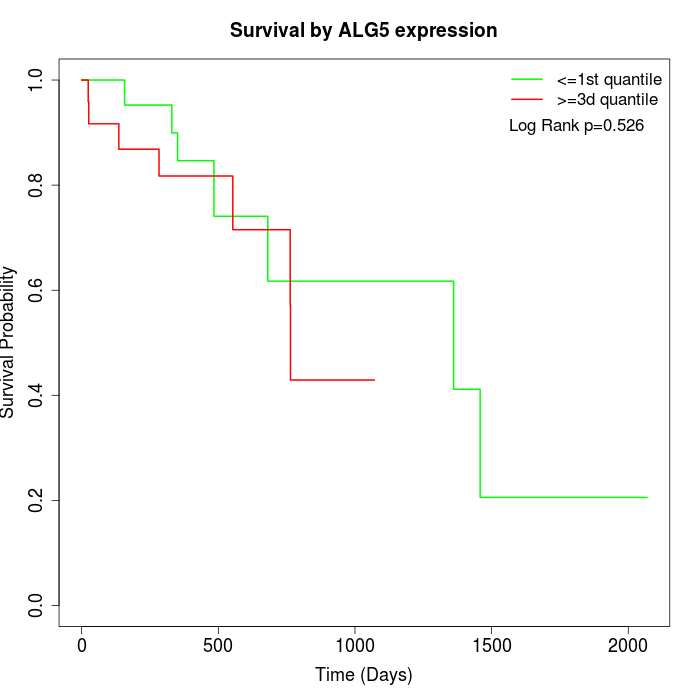
Survival by ALG5 expression:
Note: Click image to view full size file.
Copy number change of ALG5:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ALG5 | 29880 | 0 | 16 | 14 | |
| GSE20123 | ALG5 | 29880 | 0 | 15 | 15 | |
| GSE43470 | ALG5 | 29880 | 2 | 14 | 27 | |
| GSE46452 | ALG5 | 29880 | 0 | 33 | 26 | |
| GSE47630 | ALG5 | 29880 | 2 | 27 | 11 | |
| GSE54993 | ALG5 | 29880 | 12 | 2 | 56 | |
| GSE54994 | ALG5 | 29880 | 1 | 17 | 35 | |
| GSE60625 | ALG5 | 29880 | 0 | 3 | 8 | |
| GSE74703 | ALG5 | 29880 | 2 | 11 | 23 | |
| GSE74704 | ALG5 | 29880 | 0 | 12 | 8 | |
| TCGA | ALG5 | 29880 | 7 | 40 | 49 |
Total number of gains: 26; Total number of losses: 190; Total Number of normals: 272.
Somatic mutations of ALG5:
Generating mutation plots.
Highly correlated genes for ALG5:
Showing top 20/504 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ALG5 | CASP4 | 0.757976 | 3 | 0 | 3 |
| ALG5 | DECR1 | 0.730321 | 3 | 0 | 3 |
| ALG5 | ZNF564 | 0.727282 | 3 | 0 | 3 |
| ALG5 | TMEM167A | 0.725885 | 4 | 0 | 3 |
| ALG5 | ANKRD11 | 0.710315 | 3 | 0 | 3 |
| ALG5 | HYLS1 | 0.691705 | 3 | 0 | 3 |
| ALG5 | RNF149 | 0.690148 | 3 | 0 | 3 |
| ALG5 | GTF3C4 | 0.686363 | 3 | 0 | 3 |
| ALG5 | VPS36 | 0.685794 | 4 | 0 | 3 |
| ALG5 | GMCL1 | 0.681458 | 3 | 0 | 3 |
| ALG5 | BAG4 | 0.681109 | 4 | 0 | 3 |
| ALG5 | LATS1 | 0.679163 | 4 | 0 | 3 |
| ALG5 | DHRSX | 0.678173 | 3 | 0 | 3 |
| ALG5 | SPATA9 | 0.675924 | 3 | 0 | 3 |
| ALG5 | ENTPD4 | 0.675409 | 3 | 0 | 3 |
| ALG5 | PIGU | 0.674146 | 3 | 0 | 3 |
| ALG5 | THAP2 | 0.67393 | 3 | 0 | 3 |
| ALG5 | THAP5 | 0.672869 | 3 | 0 | 3 |
| ALG5 | UBXN2A | 0.671637 | 3 | 0 | 3 |
| ALG5 | TTC30B | 0.671386 | 4 | 0 | 3 |
For details and further investigation, click here