| Full name: large tumor suppressor kinase 1 | Alias Symbol: WARTS | ||
| Type: protein-coding gene | Cytoband: 6q25.1 | ||
| Entrez ID: 9113 | HGNC ID: HGNC:6514 | Ensembl Gene: ENSG00000131023 | OMIM ID: 603473 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
LATS1 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04390 | Hippo signaling pathway |
Expression of LATS1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LATS1 | 9113 | 227772_at | 0.1451 | 0.7140 | |
| GSE20347 | LATS1 | 9113 | 219813_at | -0.0127 | 0.8550 | |
| GSE23400 | LATS1 | 9113 | 219813_at | 0.0403 | 0.0412 | |
| GSE26886 | LATS1 | 9113 | 227772_at | 0.0649 | 0.8085 | |
| GSE29001 | LATS1 | 9113 | 219813_at | 0.0076 | 0.9606 | |
| GSE38129 | LATS1 | 9113 | 219813_at | -0.0696 | 0.0988 | |
| GSE45670 | LATS1 | 9113 | 227772_at | -0.0466 | 0.7888 | |
| GSE53622 | LATS1 | 9113 | 140106 | 0.0529 | 0.4113 | |
| GSE53624 | LATS1 | 9113 | 140106 | -0.0130 | 0.9075 | |
| GSE63941 | LATS1 | 9113 | 227772_at | -0.6485 | 0.1189 | |
| GSE77861 | LATS1 | 9113 | 227772_at | 0.3136 | 0.1888 | |
| GSE97050 | LATS1 | 9113 | A_33_P3233764 | 0.1524 | 0.5992 | |
| SRP007169 | LATS1 | 9113 | RNAseq | 0.0260 | 0.9353 | |
| SRP008496 | LATS1 | 9113 | RNAseq | -0.0300 | 0.8639 | |
| SRP064894 | LATS1 | 9113 | RNAseq | -0.0002 | 0.9994 | |
| SRP133303 | LATS1 | 9113 | RNAseq | 0.2031 | 0.1408 | |
| SRP159526 | LATS1 | 9113 | RNAseq | 0.0398 | 0.8723 | |
| SRP193095 | LATS1 | 9113 | RNAseq | 0.1172 | 0.1462 | |
| SRP219564 | LATS1 | 9113 | RNAseq | 0.0660 | 0.7991 | |
| TCGA | LATS1 | 9113 | RNAseq | -0.0314 | 0.5865 |
Upregulated datasets: 0; Downregulated datasets: 0.
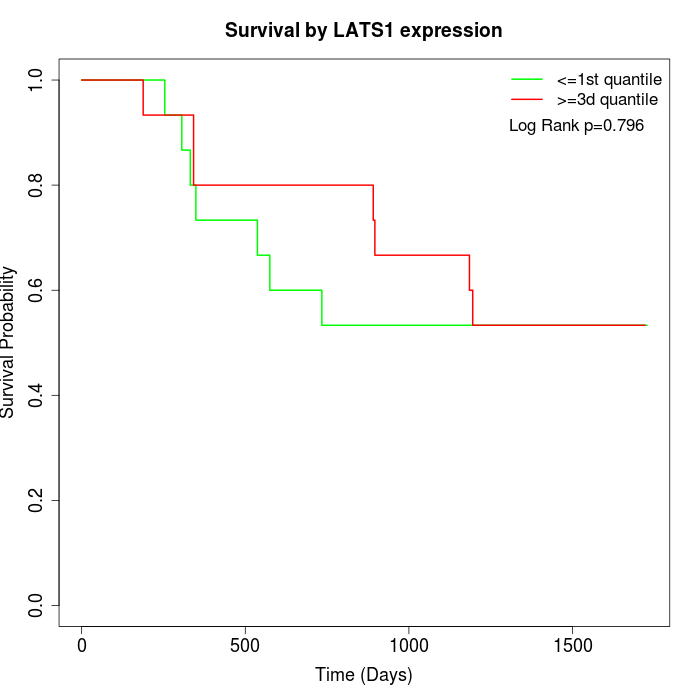
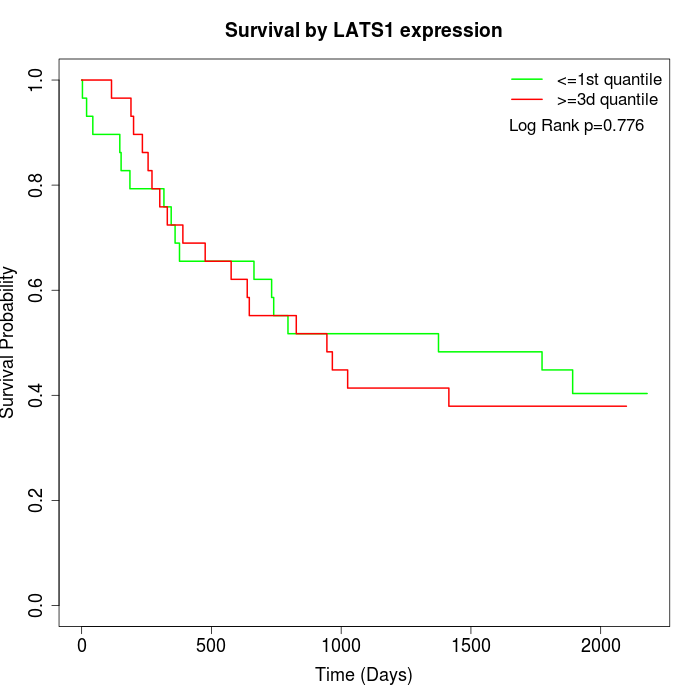
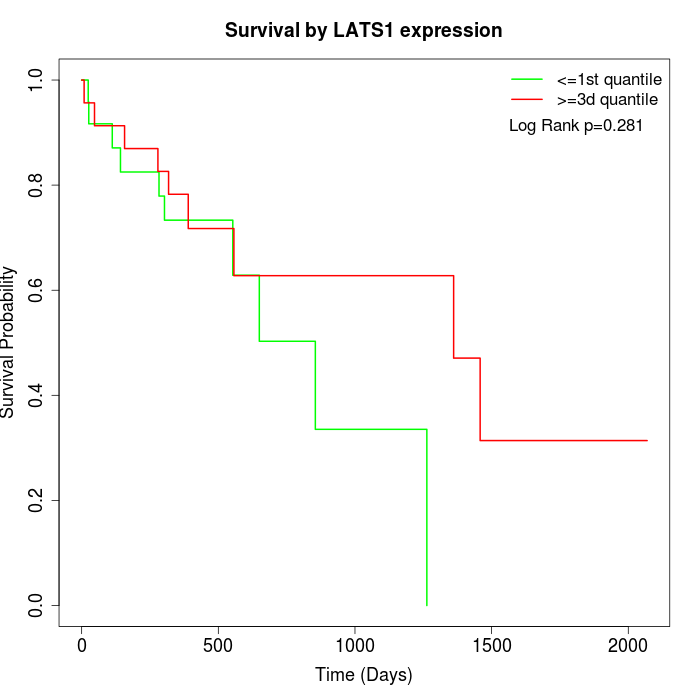
Survival by LATS1 expression:
Note: Click image to view full size file.
Copy number change of LATS1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LATS1 | 9113 | 2 | 4 | 24 | |
| GSE20123 | LATS1 | 9113 | 1 | 3 | 26 | |
| GSE43470 | LATS1 | 9113 | 4 | 0 | 39 | |
| GSE46452 | LATS1 | 9113 | 3 | 10 | 46 | |
| GSE47630 | LATS1 | 9113 | 9 | 4 | 27 | |
| GSE54993 | LATS1 | 9113 | 3 | 2 | 65 | |
| GSE54994 | LATS1 | 9113 | 8 | 8 | 37 | |
| GSE60625 | LATS1 | 9113 | 0 | 1 | 10 | |
| GSE74703 | LATS1 | 9113 | 4 | 0 | 32 | |
| GSE74704 | LATS1 | 9113 | 0 | 1 | 19 | |
| TCGA | LATS1 | 9113 | 10 | 22 | 64 |
Total number of gains: 44; Total number of losses: 55; Total Number of normals: 389.
Somatic mutations of LATS1:
Generating mutation plots.
Highly correlated genes for LATS1:
Showing top 20/288 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LATS1 | ACBD5 | 0.783571 | 3 | 0 | 3 |
| LATS1 | RBBP6 | 0.771921 | 3 | 0 | 3 |
| LATS1 | PIGM | 0.762171 | 3 | 0 | 3 |
| LATS1 | CNOT2 | 0.75448 | 3 | 0 | 3 |
| LATS1 | LRP11 | 0.747986 | 3 | 0 | 3 |
| LATS1 | NPAT | 0.747526 | 3 | 0 | 3 |
| LATS1 | MBTPS2 | 0.736888 | 4 | 0 | 4 |
| LATS1 | LARP4 | 0.733078 | 3 | 0 | 3 |
| LATS1 | C17orf80 | 0.72959 | 4 | 0 | 4 |
| LATS1 | CAND1 | 0.72352 | 3 | 0 | 3 |
| LATS1 | NEK4 | 0.720976 | 4 | 0 | 3 |
| LATS1 | SCYL3 | 0.715467 | 3 | 0 | 3 |
| LATS1 | TADA1 | 0.714649 | 4 | 0 | 4 |
| LATS1 | NAA30 | 0.713106 | 3 | 0 | 3 |
| LATS1 | CEP44 | 0.712843 | 4 | 0 | 4 |
| LATS1 | BTF3L4 | 0.711997 | 3 | 0 | 3 |
| LATS1 | KLHL8 | 0.707974 | 4 | 0 | 4 |
| LATS1 | EXOC8 | 0.705121 | 5 | 0 | 5 |
| LATS1 | ESF1 | 0.705017 | 4 | 0 | 4 |
| LATS1 | PIGB | 0.703065 | 3 | 0 | 3 |
For details and further investigation, click here