| Full name: anti-Mullerian hormone | Alias Symbol: MIS | ||
| Type: protein-coding gene | Cytoband: 19p13.3 | ||
| Entrez ID: 268 | HGNC ID: HGNC:464 | Ensembl Gene: ENSG00000104899 | OMIM ID: 600957 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
AMH involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04024 | cAMP signaling pathway | |
| hsa04350 | TGF-beta signaling pathway | |
| hsa04390 | Hippo signaling pathway |
Expression of AMH:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE53622 | AMH | 268 | 20930 | 0.0679 | 0.2975 | |
| GSE53624 | AMH | 268 | 20930 | 0.2151 | 0.0008 | |
| GSE97050 | AMH | 268 | A_23_P78944 | -0.1993 | 0.3961 | |
| SRP219564 | AMH | 268 | RNAseq | 0.2861 | 0.6250 | |
| TCGA | AMH | 268 | RNAseq | 0.6465 | 0.0967 |
Upregulated datasets: 0; Downregulated datasets: 0.
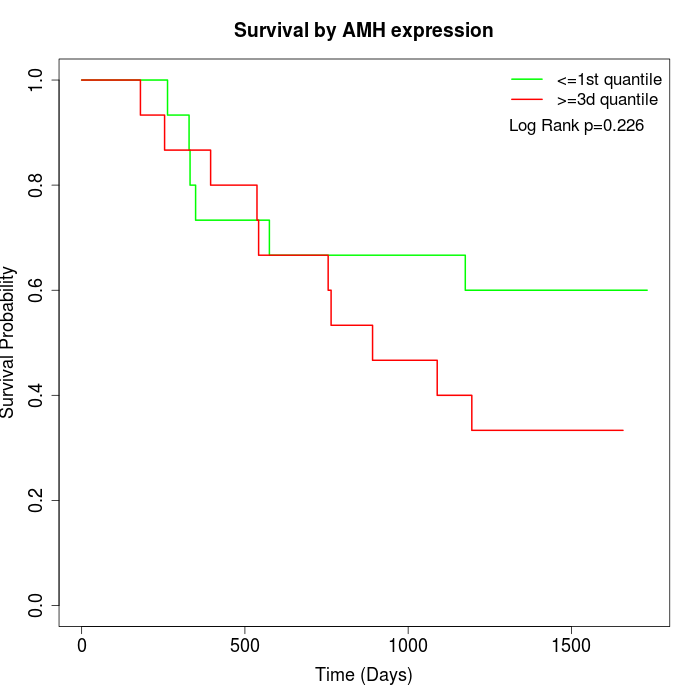
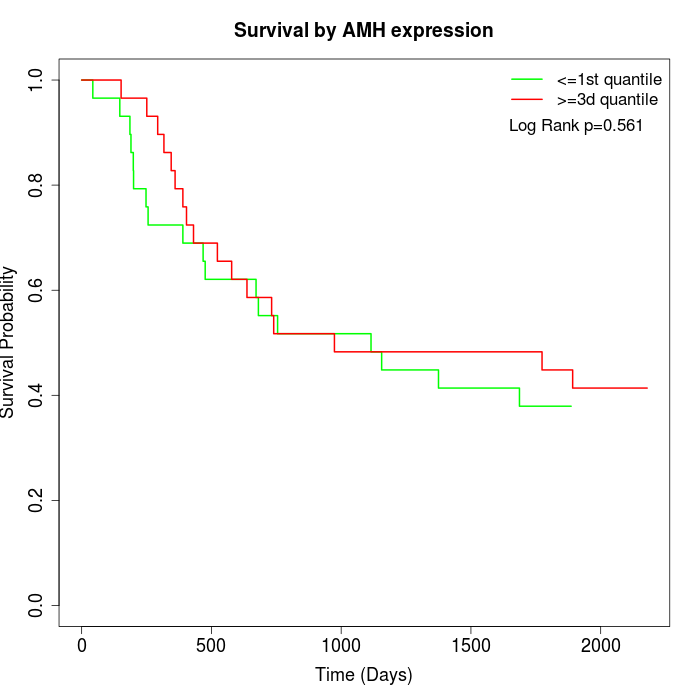
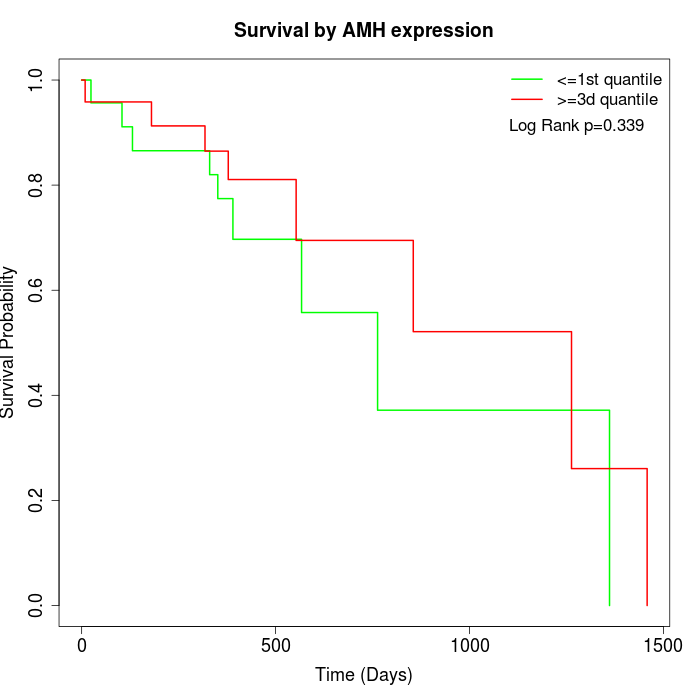
Survival by AMH expression:
Note: Click image to view full size file.
Copy number change of AMH:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | AMH | 268 | 3 | 6 | 21 | |
| GSE20123 | AMH | 268 | 4 | 4 | 22 | |
| GSE43470 | AMH | 268 | 1 | 10 | 32 | |
| GSE46452 | AMH | 268 | 47 | 1 | 11 | |
| GSE47630 | AMH | 268 | 5 | 7 | 28 | |
| GSE54993 | AMH | 268 | 16 | 3 | 51 | |
| GSE54994 | AMH | 268 | 8 | 15 | 30 | |
| GSE60625 | AMH | 268 | 9 | 0 | 2 | |
| GSE74703 | AMH | 268 | 1 | 7 | 28 | |
| GSE74704 | AMH | 268 | 2 | 3 | 15 | |
| TCGA | AMH | 268 | 7 | 24 | 65 |
Total number of gains: 103; Total number of losses: 80; Total Number of normals: 305.
Somatic mutations of AMH:
Generating mutation plots.
Highly correlated genes for AMH:
Showing top 20/30 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| AMH | NXNL2 | 0.739684 | 3 | 0 | 3 |
| AMH | SLC13A2 | 0.722809 | 3 | 0 | 3 |
| AMH | TMEM63B | 0.71989 | 3 | 0 | 3 |
| AMH | CCDC154 | 0.699703 | 3 | 0 | 3 |
| AMH | RAVER1 | 0.693976 | 3 | 0 | 3 |
| AMH | RIT2 | 0.691015 | 3 | 0 | 3 |
| AMH | OR1L6 | 0.687949 | 3 | 0 | 3 |
| AMH | RSPH9 | 0.677725 | 3 | 0 | 3 |
| AMH | SPEG | 0.670222 | 3 | 0 | 3 |
| AMH | CDH24 | 0.669001 | 3 | 0 | 3 |
| AMH | KRTAP10-3 | 0.661148 | 3 | 0 | 3 |
| AMH | HES6 | 0.660393 | 3 | 0 | 3 |
| AMH | NF2 | 0.656517 | 3 | 0 | 3 |
| AMH | C16orf90 | 0.652104 | 3 | 0 | 3 |
| AMH | LAMC3 | 0.65112 | 3 | 0 | 3 |
| AMH | KRTAP10-9 | 0.650932 | 3 | 0 | 3 |
| AMH | CNTNAP1 | 0.645206 | 3 | 0 | 3 |
| AMH | NRTN | 0.639504 | 3 | 0 | 3 |
| AMH | AVP | 0.637242 | 3 | 0 | 3 |
| AMH | ELFN1 | 0.635201 | 3 | 0 | 3 |
For details and further investigation, click here