| Full name: arginine vasopressin | Alias Symbol: ADH | ||
| Type: protein-coding gene | Cytoband: 20p13 | ||
| Entrez ID: 551 | HGNC ID: HGNC:894 | Ensembl Gene: ENSG00000101200 | OMIM ID: 192340 |
| Drug and gene relationship at DGIdb | |||
AVP involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04072 | Phospholipase D signaling pathway | |
| hsa04962 | Vasopressin-regulated water reabsorption |
Expression of AVP:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | AVP | 551 | 207848_at | -0.0096 | 0.9740 | |
| GSE20347 | AVP | 551 | 207848_at | -0.0888 | 0.1324 | |
| GSE23400 | AVP | 551 | 207848_at | -0.0754 | 0.0065 | |
| GSE26886 | AVP | 551 | 207848_at | 0.1652 | 0.1993 | |
| GSE29001 | AVP | 551 | 207848_at | -0.1325 | 0.2650 | |
| GSE38129 | AVP | 551 | 207848_at | -0.0814 | 0.2376 | |
| GSE45670 | AVP | 551 | 207848_at | -0.1075 | 0.2165 | |
| GSE53622 | AVP | 551 | 11281 | -0.1003 | 0.0433 | |
| GSE53624 | AVP | 551 | 11281 | -0.1182 | 0.0057 | |
| GSE63941 | AVP | 551 | 207848_at | 0.4145 | 0.0021 | |
| GSE77861 | AVP | 551 | 207848_at | -0.1196 | 0.1919 | |
| GSE97050 | AVP | 551 | A_23_P109133 | 0.0214 | 0.9314 |
Upregulated datasets: 0; Downregulated datasets: 0.
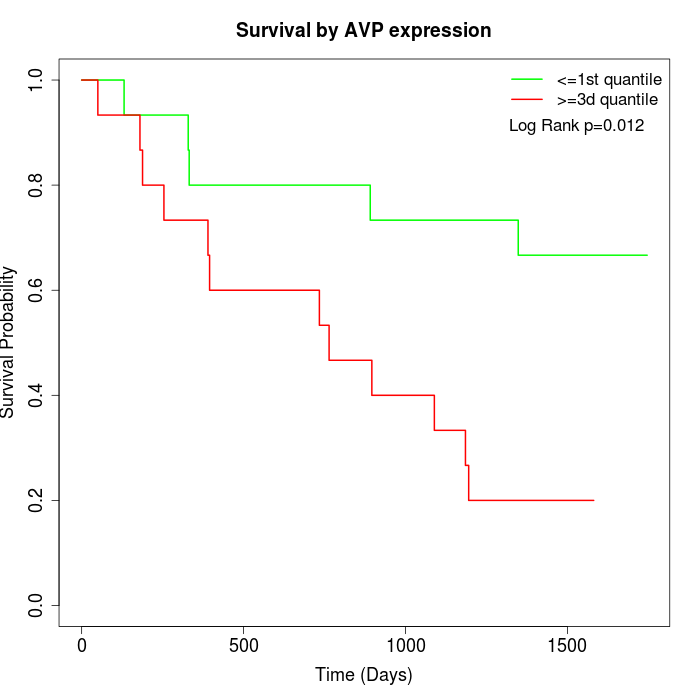
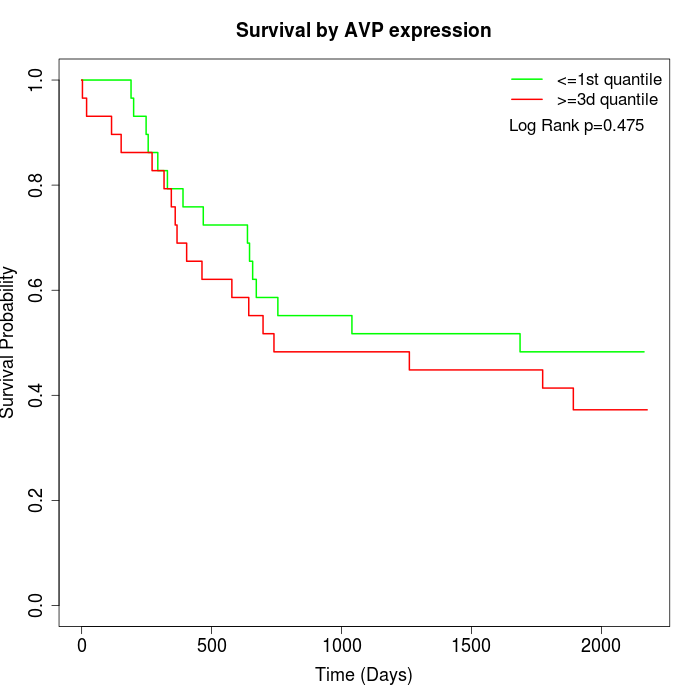
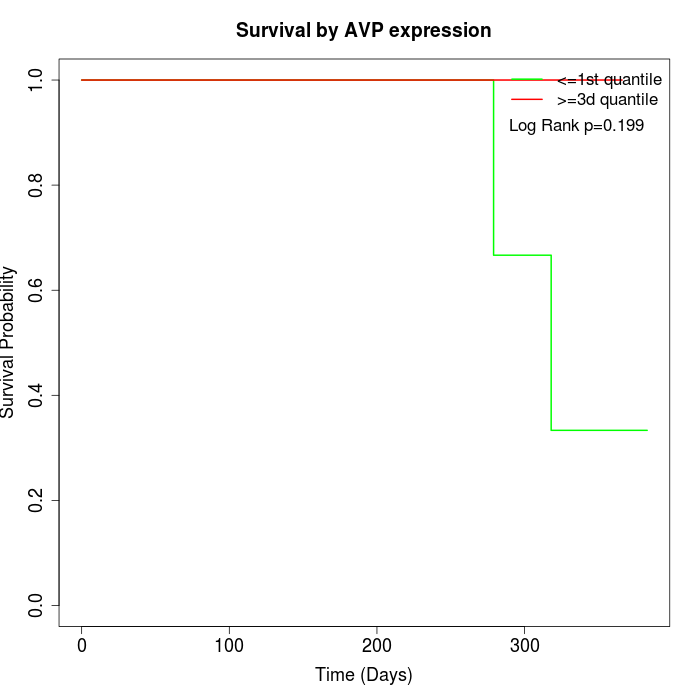
Survival by AVP expression:
Note: Click image to view full size file.
Copy number change of AVP:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | AVP | 551 | 12 | 3 | 15 | |
| GSE20123 | AVP | 551 | 13 | 3 | 14 | |
| GSE43470 | AVP | 551 | 10 | 1 | 32 | |
| GSE46452 | AVP | 551 | 26 | 1 | 32 | |
| GSE47630 | AVP | 551 | 18 | 4 | 18 | |
| GSE54993 | AVP | 551 | 2 | 14 | 54 | |
| GSE54994 | AVP | 551 | 24 | 2 | 27 | |
| GSE60625 | AVP | 551 | 0 | 0 | 11 | |
| GSE74703 | AVP | 551 | 9 | 1 | 26 | |
| GSE74704 | AVP | 551 | 8 | 1 | 11 | |
| TCGA | AVP | 551 | 40 | 11 | 45 |
Total number of gains: 162; Total number of losses: 41; Total Number of normals: 285.
Somatic mutations of AVP:
Generating mutation plots.
Highly correlated genes for AVP:
Showing top 20/549 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| AVP | KRTAP5-8 | 0.787534 | 4 | 0 | 3 |
| AVP | GUK1 | 0.783442 | 3 | 0 | 3 |
| AVP | AARS2 | 0.771818 | 3 | 0 | 3 |
| AVP | CLDND2 | 0.765301 | 3 | 0 | 3 |
| AVP | C3orf80 | 0.735941 | 3 | 0 | 3 |
| AVP | OR2F1 | 0.723147 | 3 | 0 | 3 |
| AVP | PRRC2A | 0.720227 | 4 | 0 | 4 |
| AVP | KCNK2 | 0.718313 | 4 | 0 | 4 |
| AVP | ARHGEF15 | 0.713626 | 5 | 0 | 4 |
| AVP | ATF7 | 0.710481 | 4 | 0 | 4 |
| AVP | NPFF | 0.708862 | 3 | 0 | 3 |
| AVP | EGLN2 | 0.701118 | 4 | 0 | 3 |
| AVP | BTBD2 | 0.700722 | 3 | 0 | 3 |
| AVP | FRAS1 | 0.698693 | 3 | 0 | 3 |
| AVP | MS4A1 | 0.697619 | 3 | 0 | 3 |
| AVP | CYP2C19 | 0.696744 | 3 | 0 | 3 |
| AVP | EDA2R | 0.696108 | 4 | 0 | 4 |
| AVP | CYP3A4 | 0.690018 | 3 | 0 | 3 |
| AVP | OTUD7B | 0.689452 | 3 | 0 | 3 |
| AVP | PPP2R1A | 0.68895 | 4 | 0 | 3 |
For details and further investigation, click here