| Full name: adaptor related protein complex 3 subunit mu 2 | Alias Symbol: CLA20|AP47B | ||
| Type: protein-coding gene | Cytoband: 8p11.21 | ||
| Entrez ID: 10947 | HGNC ID: HGNC:570 | Ensembl Gene: ENSG00000070718 | OMIM ID: 610469 |
| Drug and gene relationship at DGIdb | |||
Expression of AP3M2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | AP3M2 | 10947 | 203410_at | 0.5998 | 0.2698 | |
| GSE20347 | AP3M2 | 10947 | 203410_at | 1.0719 | 0.0000 | |
| GSE23400 | AP3M2 | 10947 | 203410_at | 0.5440 | 0.0000 | |
| GSE26886 | AP3M2 | 10947 | 203410_at | 0.5677 | 0.0201 | |
| GSE29001 | AP3M2 | 10947 | 203410_at | 0.9186 | 0.0016 | |
| GSE38129 | AP3M2 | 10947 | 203410_at | 0.8352 | 0.0000 | |
| GSE45670 | AP3M2 | 10947 | 203410_at | 0.3922 | 0.0159 | |
| GSE53622 | AP3M2 | 10947 | 816 | 0.6721 | 0.0000 | |
| GSE53624 | AP3M2 | 10947 | 816 | 1.0198 | 0.0000 | |
| GSE63941 | AP3M2 | 10947 | 203410_at | -0.5085 | 0.4528 | |
| GSE77861 | AP3M2 | 10947 | 1569053_at | 0.2776 | 0.3020 | |
| GSE97050 | AP3M2 | 10947 | A_24_P64039 | 0.3149 | 0.3225 | |
| SRP007169 | AP3M2 | 10947 | RNAseq | 1.8550 | 0.0001 | |
| SRP008496 | AP3M2 | 10947 | RNAseq | 1.5625 | 0.0000 | |
| SRP064894 | AP3M2 | 10947 | RNAseq | 1.0142 | 0.0000 | |
| SRP133303 | AP3M2 | 10947 | RNAseq | 0.7459 | 0.0001 | |
| SRP159526 | AP3M2 | 10947 | RNAseq | 0.8965 | 0.0000 | |
| SRP193095 | AP3M2 | 10947 | RNAseq | 0.8561 | 0.0000 | |
| SRP219564 | AP3M2 | 10947 | RNAseq | 0.7008 | 0.0134 | |
| TCGA | AP3M2 | 10947 | RNAseq | 0.3271 | 0.0000 |
Upregulated datasets: 5; Downregulated datasets: 0.
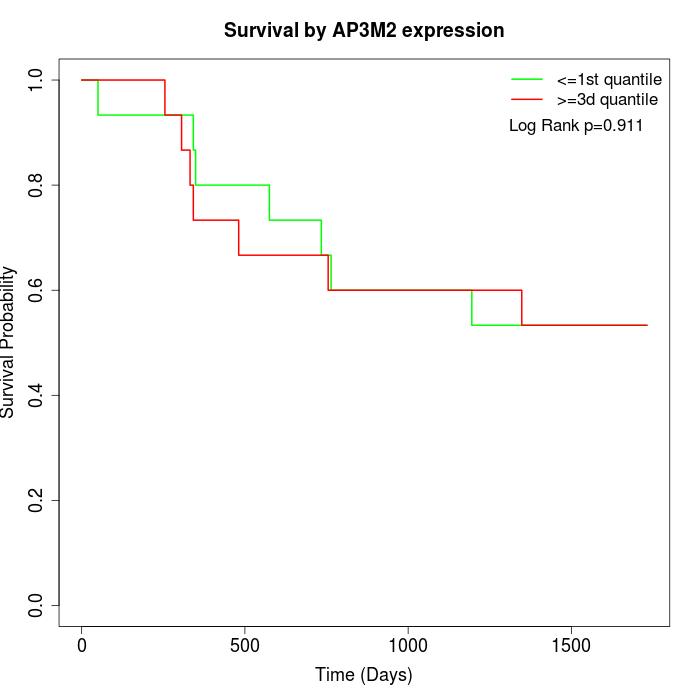
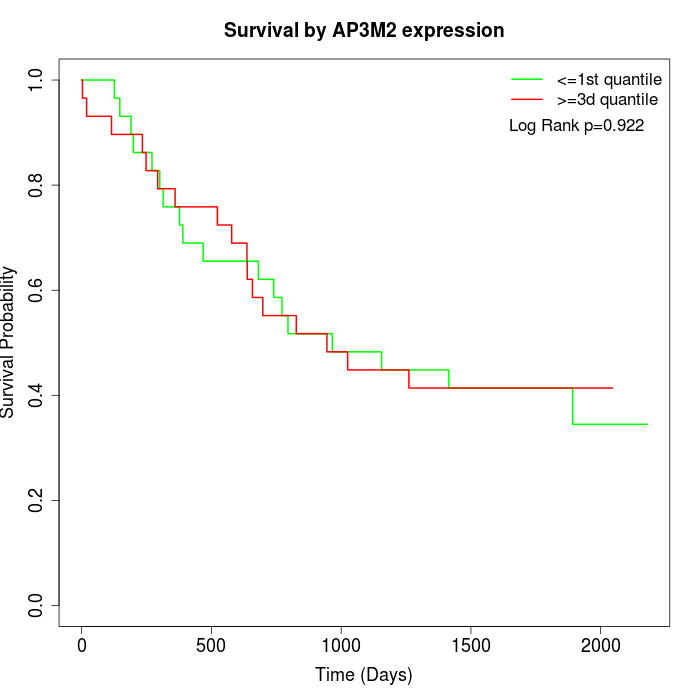
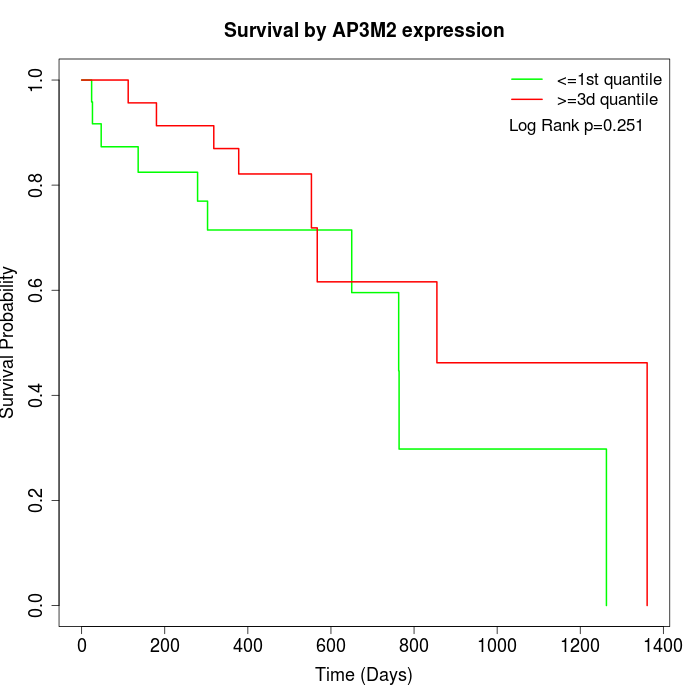
Survival by AP3M2 expression:
Note: Click image to view full size file.
Copy number change of AP3M2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | AP3M2 | 10947 | 10 | 5 | 15 | |
| GSE20123 | AP3M2 | 10947 | 10 | 5 | 15 | |
| GSE43470 | AP3M2 | 10947 | 9 | 6 | 28 | |
| GSE46452 | AP3M2 | 10947 | 19 | 5 | 35 | |
| GSE47630 | AP3M2 | 10947 | 22 | 1 | 17 | |
| GSE54993 | AP3M2 | 10947 | 1 | 16 | 53 | |
| GSE54994 | AP3M2 | 10947 | 19 | 7 | 27 | |
| GSE60625 | AP3M2 | 10947 | 3 | 0 | 8 | |
| GSE74703 | AP3M2 | 10947 | 8 | 5 | 23 | |
| GSE74704 | AP3M2 | 10947 | 9 | 2 | 9 | |
| TCGA | AP3M2 | 10947 | 40 | 22 | 34 |
Total number of gains: 150; Total number of losses: 74; Total Number of normals: 264.
Somatic mutations of AP3M2:
Generating mutation plots.
Highly correlated genes for AP3M2:
Showing top 20/1603 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| AP3M2 | SBNO1 | 0.81512 | 3 | 0 | 3 |
| AP3M2 | PEG10 | 0.790658 | 3 | 0 | 3 |
| AP3M2 | ISY1 | 0.784366 | 3 | 0 | 3 |
| AP3M2 | BRDT | 0.766105 | 3 | 0 | 3 |
| AP3M2 | ELOF1 | 0.759888 | 3 | 0 | 3 |
| AP3M2 | GRB2 | 0.759369 | 3 | 0 | 3 |
| AP3M2 | NAIF1 | 0.738696 | 3 | 0 | 3 |
| AP3M2 | G3BP2 | 0.730734 | 3 | 0 | 3 |
| AP3M2 | MSH6 | 0.728773 | 11 | 0 | 11 |
| AP3M2 | TRIM44 | 0.716271 | 3 | 0 | 3 |
| AP3M2 | TUBE1 | 0.71576 | 3 | 0 | 3 |
| AP3M2 | RBM12B | 0.715495 | 6 | 0 | 6 |
| AP3M2 | SSBP4 | 0.710689 | 4 | 0 | 4 |
| AP3M2 | C5orf51 | 0.708431 | 3 | 0 | 3 |
| AP3M2 | WDR35 | 0.706351 | 4 | 0 | 4 |
| AP3M2 | PHF3 | 0.703728 | 3 | 0 | 3 |
| AP3M2 | PIK3CB | 0.70342 | 3 | 0 | 3 |
| AP3M2 | HPS3 | 0.701643 | 5 | 0 | 4 |
| AP3M2 | RICTOR | 0.69282 | 4 | 0 | 3 |
| AP3M2 | ARHGAP18 | 0.692638 | 3 | 0 | 3 |
For details and further investigation, click here