| Full name: tripartite motif containing 44 | Alias Symbol: DIPB|MC7 | ||
| Type: protein-coding gene | Cytoband: 11p13 | ||
| Entrez ID: 54765 | HGNC ID: HGNC:19016 | Ensembl Gene: ENSG00000166326 | OMIM ID: 612298 |
| Drug and gene relationship at DGIdb | |||
Expression of TRIM44:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRIM44 | 54765 | 217759_at | -0.1435 | 0.7322 | |
| GSE20347 | TRIM44 | 54765 | 217759_at | -0.0911 | 0.6115 | |
| GSE23400 | TRIM44 | 54765 | 217759_at | 0.1356 | 0.1224 | |
| GSE26886 | TRIM44 | 54765 | 217759_at | 0.3234 | 0.2405 | |
| GSE29001 | TRIM44 | 54765 | 217759_at | 0.3400 | 0.2855 | |
| GSE38129 | TRIM44 | 54765 | 217759_at | -0.1940 | 0.2381 | |
| GSE45670 | TRIM44 | 54765 | 217759_at | -0.3103 | 0.0608 | |
| GSE53622 | TRIM44 | 54765 | 34606 | 0.0770 | 0.3526 | |
| GSE53624 | TRIM44 | 54765 | 8215 | 0.1084 | 0.1262 | |
| GSE63941 | TRIM44 | 54765 | 217759_at | -0.7901 | 0.0914 | |
| GSE77861 | TRIM44 | 54765 | 217759_at | 0.0806 | 0.8268 | |
| GSE97050 | TRIM44 | 54765 | A_23_P139297 | 0.0051 | 0.9829 | |
| SRP007169 | TRIM44 | 54765 | RNAseq | 1.3621 | 0.0007 | |
| SRP008496 | TRIM44 | 54765 | RNAseq | 1.6696 | 0.0000 | |
| SRP064894 | TRIM44 | 54765 | RNAseq | 0.8310 | 0.0032 | |
| SRP133303 | TRIM44 | 54765 | RNAseq | 0.2369 | 0.1950 | |
| SRP159526 | TRIM44 | 54765 | RNAseq | -0.0525 | 0.7581 | |
| SRP193095 | TRIM44 | 54765 | RNAseq | 0.2054 | 0.0404 | |
| SRP219564 | TRIM44 | 54765 | RNAseq | 0.1011 | 0.7382 | |
| TCGA | TRIM44 | 54765 | RNAseq | -0.1502 | 0.0043 |
Upregulated datasets: 2; Downregulated datasets: 0.
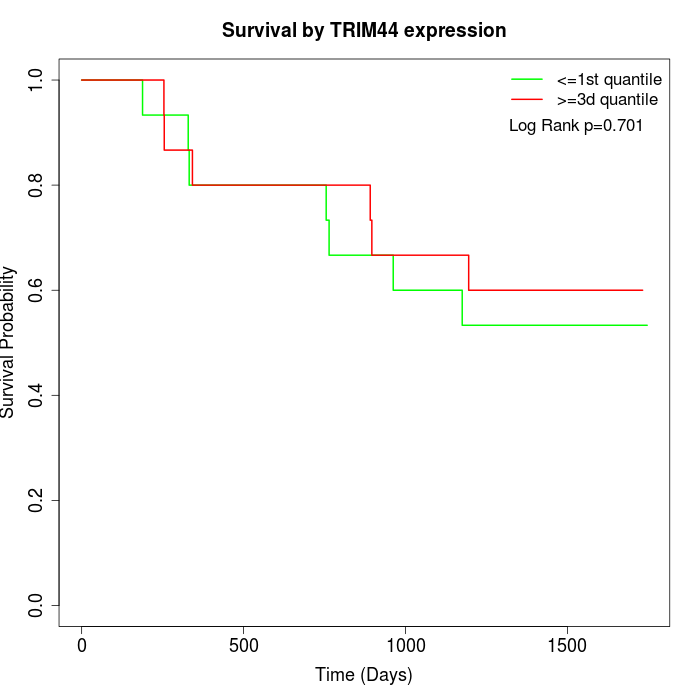
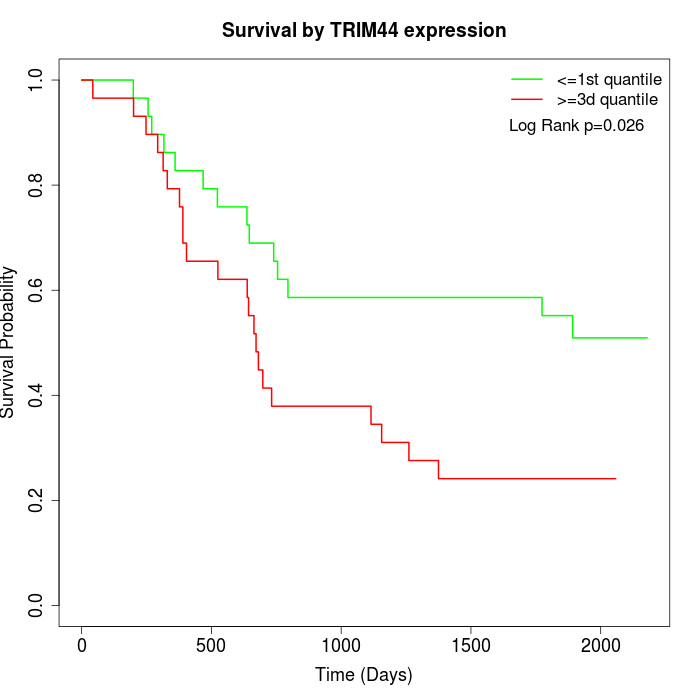
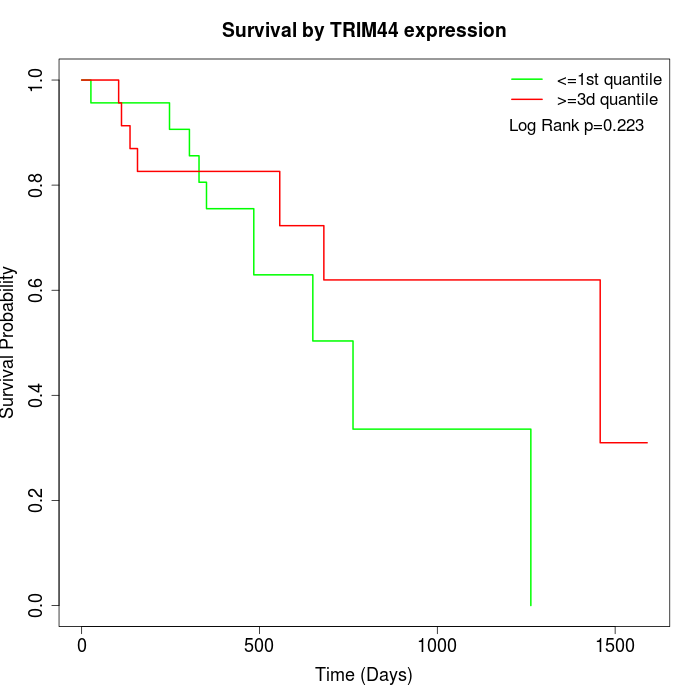
Survival by TRIM44 expression:
Note: Click image to view full size file.
Copy number change of TRIM44:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRIM44 | 54765 | 1 | 7 | 22 | |
| GSE20123 | TRIM44 | 54765 | 1 | 6 | 23 | |
| GSE43470 | TRIM44 | 54765 | 3 | 4 | 36 | |
| GSE46452 | TRIM44 | 54765 | 7 | 5 | 47 | |
| GSE47630 | TRIM44 | 54765 | 3 | 10 | 27 | |
| GSE54993 | TRIM44 | 54765 | 3 | 2 | 65 | |
| GSE54994 | TRIM44 | 54765 | 3 | 10 | 40 | |
| GSE60625 | TRIM44 | 54765 | 0 | 0 | 11 | |
| GSE74703 | TRIM44 | 54765 | 2 | 2 | 32 | |
| GSE74704 | TRIM44 | 54765 | 0 | 4 | 16 | |
| TCGA | TRIM44 | 54765 | 13 | 23 | 60 |
Total number of gains: 36; Total number of losses: 73; Total Number of normals: 379.
Somatic mutations of TRIM44:
Generating mutation plots.
Highly correlated genes for TRIM44:
Showing top 20/602 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRIM44 | COPS2 | 0.768207 | 3 | 0 | 3 |
| TRIM44 | SBF2 | 0.752869 | 4 | 0 | 4 |
| TRIM44 | AKAP7 | 0.750646 | 3 | 0 | 3 |
| TRIM44 | C20orf194 | 0.749101 | 3 | 0 | 3 |
| TRIM44 | POGK | 0.729232 | 3 | 0 | 3 |
| TRIM44 | AIMP1 | 0.7201 | 4 | 0 | 4 |
| TRIM44 | AP3M2 | 0.716271 | 3 | 0 | 3 |
| TRIM44 | PGRMC1 | 0.715729 | 4 | 0 | 4 |
| TRIM44 | ORAI3 | 0.713845 | 3 | 0 | 3 |
| TRIM44 | S100B | 0.71348 | 3 | 0 | 3 |
| TRIM44 | MRPL24 | 0.707544 | 3 | 0 | 3 |
| TRIM44 | ZNF518B | 0.699246 | 3 | 0 | 3 |
| TRIM44 | HERPUD1 | 0.693869 | 3 | 0 | 3 |
| TRIM44 | GBA2 | 0.69367 | 3 | 0 | 3 |
| TRIM44 | HAUS1 | 0.6932 | 3 | 0 | 3 |
| TRIM44 | PTPMT1 | 0.690754 | 3 | 0 | 3 |
| TRIM44 | SCML1 | 0.689534 | 4 | 0 | 3 |
| TRIM44 | SLFN11 | 0.689045 | 3 | 0 | 3 |
| TRIM44 | BEND5 | 0.687509 | 4 | 0 | 3 |
| TRIM44 | GLUL | 0.680425 | 3 | 0 | 3 |
For details and further investigation, click here