| Full name: phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 3q22.3 | ||
| Entrez ID: 5291 | HGNC ID: HGNC:8976 | Ensembl Gene: ENSG00000051382 | OMIM ID: 602925 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
PIK3CB involved pathways:
Expression of PIK3CB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | PIK3CB | 5291 | 212688_at | -0.0466 | 0.9508 | |
| GSE20347 | PIK3CB | 5291 | 212688_at | -0.1268 | 0.6069 | |
| GSE23400 | PIK3CB | 5291 | 212688_at | 0.0397 | 0.6917 | |
| GSE26886 | PIK3CB | 5291 | 212688_at | -0.4193 | 0.0723 | |
| GSE29001 | PIK3CB | 5291 | 212688_at | 0.0556 | 0.8811 | |
| GSE38129 | PIK3CB | 5291 | 212688_at | -0.1535 | 0.3970 | |
| GSE45670 | PIK3CB | 5291 | 212688_at | -0.0278 | 0.9009 | |
| GSE53622 | PIK3CB | 5291 | 47856 | 0.0582 | 0.5566 | |
| GSE53624 | PIK3CB | 5291 | 47856 | -0.0267 | 0.6752 | |
| GSE63941 | PIK3CB | 5291 | 212688_at | 0.9796 | 0.0080 | |
| GSE77861 | PIK3CB | 5291 | 212688_at | -0.5064 | 0.1250 | |
| GSE97050 | PIK3CB | 5291 | A_23_P346969 | 0.4162 | 0.2151 | |
| SRP007169 | PIK3CB | 5291 | RNAseq | 0.5947 | 0.1463 | |
| SRP008496 | PIK3CB | 5291 | RNAseq | 0.3116 | 0.2249 | |
| SRP064894 | PIK3CB | 5291 | RNAseq | -0.2025 | 0.2428 | |
| SRP133303 | PIK3CB | 5291 | RNAseq | 0.1814 | 0.1459 | |
| SRP159526 | PIK3CB | 5291 | RNAseq | 0.2378 | 0.5172 | |
| SRP193095 | PIK3CB | 5291 | RNAseq | -0.0690 | 0.5125 | |
| SRP219564 | PIK3CB | 5291 | RNAseq | -0.3750 | 0.1731 | |
| TCGA | PIK3CB | 5291 | RNAseq | 0.0070 | 0.9022 |
Upregulated datasets: 0; Downregulated datasets: 0.
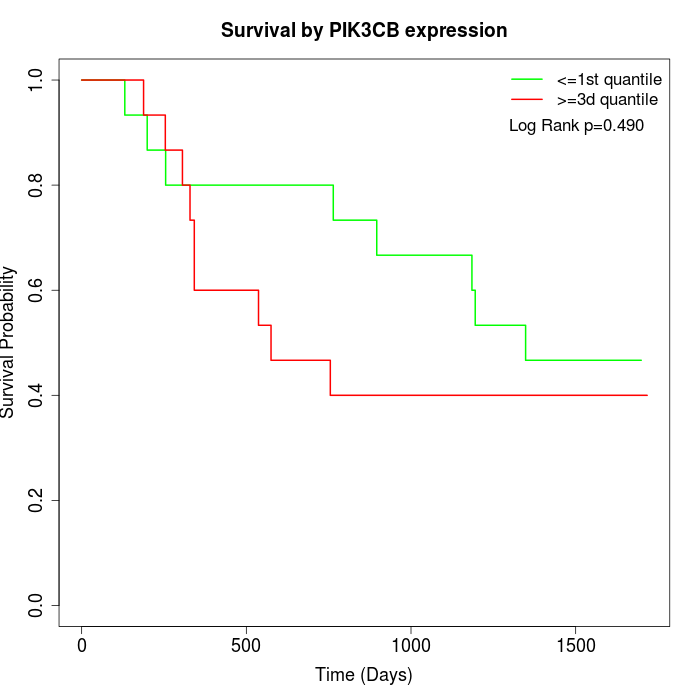
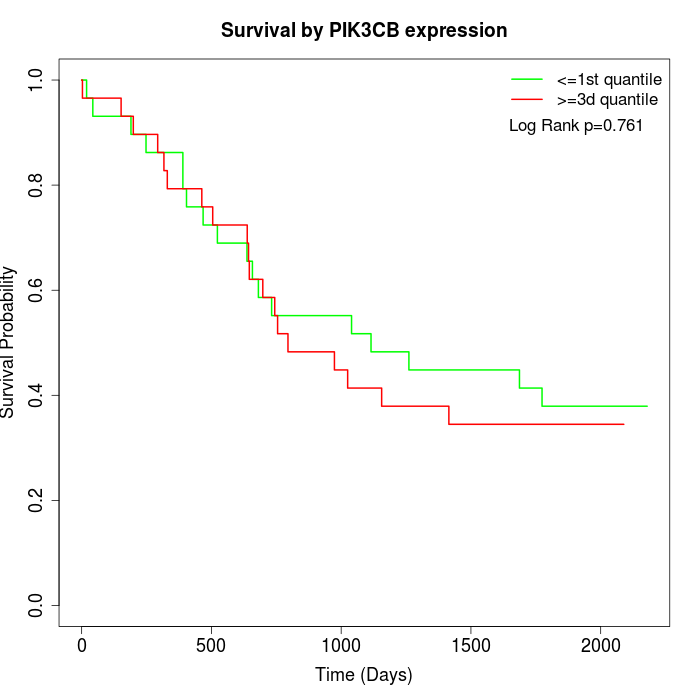
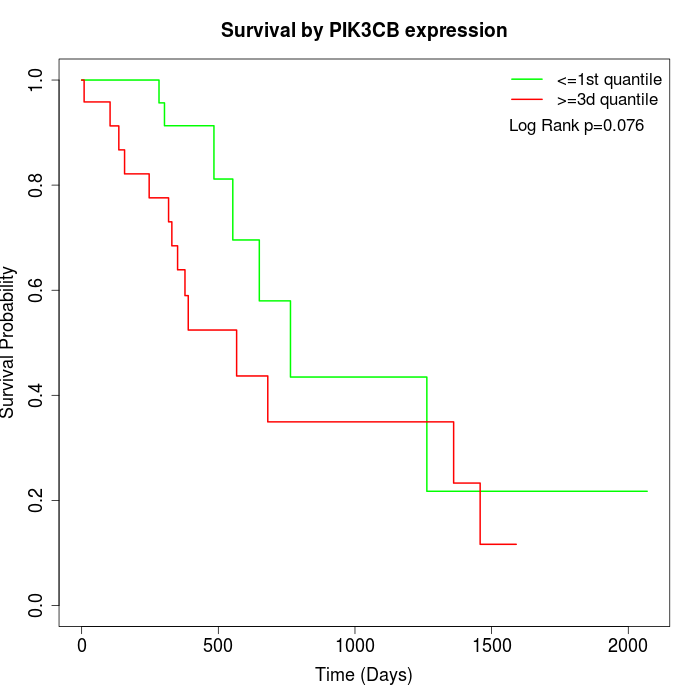
Survival by PIK3CB expression:
Note: Click image to view full size file.
Copy number change of PIK3CB:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | PIK3CB | 5291 | 17 | 0 | 13 | |
| GSE20123 | PIK3CB | 5291 | 17 | 0 | 13 | |
| GSE43470 | PIK3CB | 5291 | 22 | 0 | 21 | |
| GSE46452 | PIK3CB | 5291 | 15 | 4 | 40 | |
| GSE47630 | PIK3CB | 5291 | 18 | 3 | 19 | |
| GSE54993 | PIK3CB | 5291 | 2 | 8 | 60 | |
| GSE54994 | PIK3CB | 5291 | 32 | 1 | 20 | |
| GSE60625 | PIK3CB | 5291 | 0 | 6 | 5 | |
| GSE74703 | PIK3CB | 5291 | 19 | 0 | 17 | |
| GSE74704 | PIK3CB | 5291 | 11 | 0 | 9 | |
| TCGA | PIK3CB | 5291 | 62 | 4 | 30 |
Total number of gains: 215; Total number of losses: 26; Total Number of normals: 247.
Somatic mutations of PIK3CB:
Generating mutation plots.
Highly correlated genes for PIK3CB:
Showing top 20/431 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| PIK3CB | CSTF2T | 0.827609 | 3 | 0 | 3 |
| PIK3CB | TMEM120A | 0.807836 | 3 | 0 | 3 |
| PIK3CB | TMBIM6 | 0.794423 | 3 | 0 | 3 |
| PIK3CB | LCOR | 0.763106 | 3 | 0 | 3 |
| PIK3CB | PANK1 | 0.76054 | 3 | 0 | 3 |
| PIK3CB | NFKB1 | 0.753114 | 3 | 0 | 3 |
| PIK3CB | GLUD1 | 0.746465 | 3 | 0 | 3 |
| PIK3CB | RHBDD1 | 0.744983 | 3 | 0 | 3 |
| PIK3CB | ARL6 | 0.736734 | 4 | 0 | 4 |
| PIK3CB | C2CD2 | 0.735132 | 4 | 0 | 4 |
| PIK3CB | WDR77 | 0.734249 | 4 | 0 | 4 |
| PIK3CB | ELF2 | 0.73008 | 4 | 0 | 4 |
| PIK3CB | LRRC47 | 0.728354 | 4 | 0 | 3 |
| PIK3CB | THAP1 | 0.727212 | 3 | 0 | 3 |
| PIK3CB | THAP2 | 0.723412 | 3 | 0 | 3 |
| PIK3CB | ATG4C | 0.722898 | 3 | 0 | 3 |
| PIK3CB | EIF2B1 | 0.722331 | 4 | 0 | 3 |
| PIK3CB | MRPS18A | 0.720995 | 3 | 0 | 3 |
| PIK3CB | ZBED5 | 0.718415 | 3 | 0 | 3 |
| PIK3CB | RBM4 | 0.717872 | 3 | 0 | 3 |
For details and further investigation, click here