| Full name: Rho GDP dissociation inhibitor beta | Alias Symbol: Ly-GDI|RhoGDI2 | ||
| Type: protein-coding gene | Cytoband: 12p12.3 | ||
| Entrez ID: 397 | HGNC ID: HGNC:679 | Ensembl Gene: ENSG00000111348 | OMIM ID: 602843 |
| Drug and gene relationship at DGIdb | |||
ARHGDIB involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04722 | Neurotrophin signaling pathway | |
| hsa04962 | Vasopressin-regulated water reabsorption |
Expression of ARHGDIB:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ARHGDIB | 397 | 201288_at | 0.4330 | 0.5715 | |
| GSE20347 | ARHGDIB | 397 | 201288_at | -0.2665 | 0.2515 | |
| GSE23400 | ARHGDIB | 397 | 201288_at | 0.0373 | 0.7532 | |
| GSE26886 | ARHGDIB | 397 | 201288_at | -0.4308 | 0.2338 | |
| GSE29001 | ARHGDIB | 397 | 201288_at | 0.0255 | 0.9157 | |
| GSE38129 | ARHGDIB | 397 | 201288_at | -0.2779 | 0.2354 | |
| GSE45670 | ARHGDIB | 397 | 201288_at | -0.1260 | 0.6394 | |
| GSE53622 | ARHGDIB | 397 | 34754 | 0.0297 | 0.8079 | |
| GSE53624 | ARHGDIB | 397 | 34754 | -0.1437 | 0.0920 | |
| GSE63941 | ARHGDIB | 397 | 201288_at | 2.8955 | 0.2172 | |
| GSE77861 | ARHGDIB | 397 | 201288_at | -0.1754 | 0.6147 | |
| GSE97050 | ARHGDIB | 397 | A_23_P151075 | 0.4240 | 0.3868 | |
| SRP007169 | ARHGDIB | 397 | RNAseq | 0.4510 | 0.5056 | |
| SRP008496 | ARHGDIB | 397 | RNAseq | 0.1526 | 0.6936 | |
| SRP064894 | ARHGDIB | 397 | RNAseq | 0.3301 | 0.1728 | |
| SRP133303 | ARHGDIB | 397 | RNAseq | -0.1085 | 0.6220 | |
| SRP159526 | ARHGDIB | 397 | RNAseq | -0.7111 | 0.2218 | |
| SRP193095 | ARHGDIB | 397 | RNAseq | -0.0209 | 0.9056 | |
| SRP219564 | ARHGDIB | 397 | RNAseq | 0.4846 | 0.2143 | |
| TCGA | ARHGDIB | 397 | RNAseq | 0.1868 | 0.0303 |
Upregulated datasets: 0; Downregulated datasets: 0.
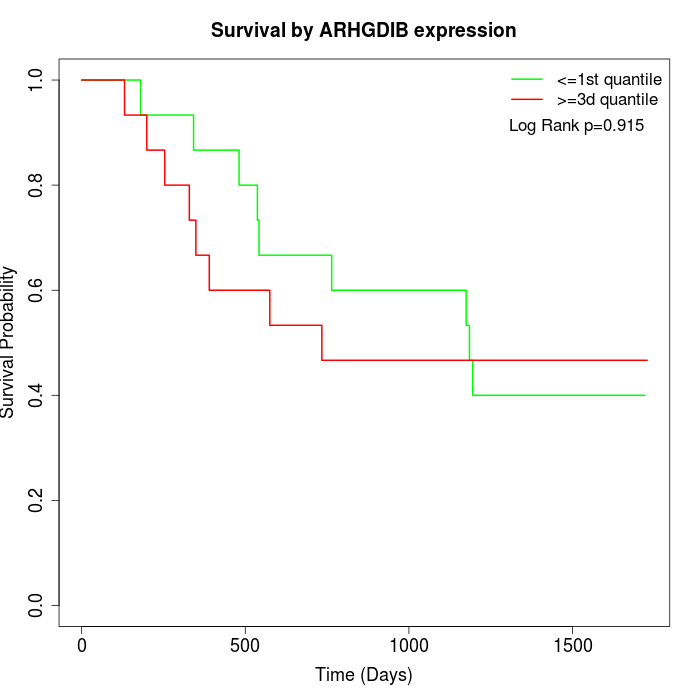
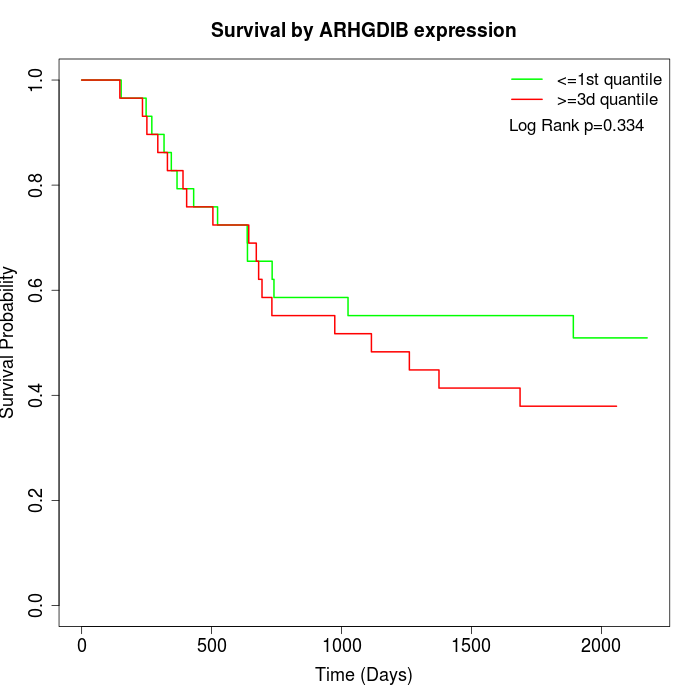
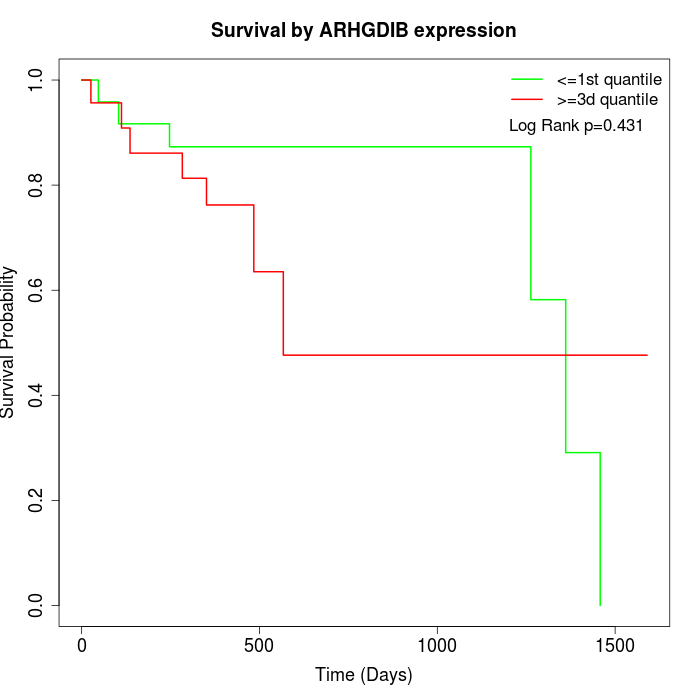
Survival by ARHGDIB expression:
Note: Click image to view full size file.
Copy number change of ARHGDIB:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ARHGDIB | 397 | 6 | 2 | 22 | |
| GSE20123 | ARHGDIB | 397 | 6 | 2 | 22 | |
| GSE43470 | ARHGDIB | 397 | 9 | 4 | 30 | |
| GSE46452 | ARHGDIB | 397 | 10 | 1 | 48 | |
| GSE47630 | ARHGDIB | 397 | 14 | 1 | 25 | |
| GSE54993 | ARHGDIB | 397 | 1 | 9 | 60 | |
| GSE54994 | ARHGDIB | 397 | 10 | 2 | 41 | |
| GSE60625 | ARHGDIB | 397 | 0 | 1 | 10 | |
| GSE74703 | ARHGDIB | 397 | 9 | 3 | 24 | |
| GSE74704 | ARHGDIB | 397 | 4 | 1 | 15 | |
| TCGA | ARHGDIB | 397 | 41 | 5 | 50 |
Total number of gains: 110; Total number of losses: 31; Total Number of normals: 347.
Somatic mutations of ARHGDIB:
Generating mutation plots.
Highly correlated genes for ARHGDIB:
Showing top 20/406 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ARHGDIB | GMFG | 0.78022 | 10 | 0 | 10 |
| ARHGDIB | SNX20 | 0.763315 | 6 | 0 | 5 |
| ARHGDIB | MPEG1 | 0.753073 | 6 | 0 | 6 |
| ARHGDIB | LAPTM5 | 0.752814 | 10 | 0 | 10 |
| ARHGDIB | TNFAIP8L2 | 0.752197 | 6 | 0 | 6 |
| ARHGDIB | IRF8 | 0.748259 | 10 | 0 | 10 |
| ARHGDIB | CORO1A | 0.745725 | 12 | 0 | 10 |
| ARHGDIB | DOCK2 | 0.745704 | 9 | 0 | 9 |
| ARHGDIB | CD53 | 0.744852 | 9 | 0 | 8 |
| ARHGDIB | CD48 | 0.744846 | 9 | 0 | 9 |
| ARHGDIB | HLA-DOA | 0.744756 | 5 | 0 | 5 |
| ARHGDIB | PTPRC | 0.739762 | 9 | 0 | 9 |
| ARHGDIB | PIK3R5 | 0.737981 | 5 | 0 | 5 |
| ARHGDIB | SLA | 0.737958 | 10 | 0 | 10 |
| ARHGDIB | ST8SIA4 | 0.73341 | 6 | 0 | 6 |
| ARHGDIB | ARHGAP9 | 0.730697 | 7 | 0 | 7 |
| ARHGDIB | IL10RA | 0.729849 | 9 | 0 | 9 |
| ARHGDIB | C16orf54 | 0.729583 | 6 | 0 | 5 |
| ARHGDIB | HLA-DMB | 0.728593 | 11 | 0 | 10 |
| ARHGDIB | C1orf162 | 0.728351 | 7 | 0 | 7 |
For details and further investigation, click here