| Full name: ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 | Alias Symbol: KIAA0400|PAP|SHAG1|CENTB3 | ||
| Type: protein-coding gene | Cytoband: 2p25.1 | ||
| Entrez ID: 8853 | HGNC ID: HGNC:2721 | Ensembl Gene: ENSG00000151693 | OMIM ID: 603817 |
| Drug and gene relationship at DGIdb | |||
ASAP2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04666 | Fc gamma R-mediated phagocytosis |
Expression of ASAP2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ASAP2 | 8853 | 206414_s_at | 0.1892 | 0.8307 | |
| GSE20347 | ASAP2 | 8853 | 206414_s_at | 0.6708 | 0.0004 | |
| GSE23400 | ASAP2 | 8853 | 206414_s_at | 0.4955 | 0.0000 | |
| GSE26886 | ASAP2 | 8853 | 206414_s_at | 0.8282 | 0.0011 | |
| GSE29001 | ASAP2 | 8853 | 206414_s_at | 0.5808 | 0.0408 | |
| GSE38129 | ASAP2 | 8853 | 206414_s_at | 0.4776 | 0.0026 | |
| GSE45670 | ASAP2 | 8853 | 206414_s_at | 0.3358 | 0.2587 | |
| GSE63941 | ASAP2 | 8853 | 206414_s_at | -0.3715 | 0.4083 | |
| GSE77861 | ASAP2 | 8853 | 206414_s_at | 1.0090 | 0.0172 | |
| GSE97050 | ASAP2 | 8853 | A_24_P362540 | 0.4363 | 0.1456 | |
| SRP007169 | ASAP2 | 8853 | RNAseq | 0.0578 | 0.9196 | |
| SRP008496 | ASAP2 | 8853 | RNAseq | 0.4511 | 0.1466 | |
| SRP064894 | ASAP2 | 8853 | RNAseq | -0.1964 | 0.6061 | |
| SRP133303 | ASAP2 | 8853 | RNAseq | 0.5199 | 0.0016 | |
| SRP159526 | ASAP2 | 8853 | RNAseq | 0.7216 | 0.0111 | |
| SRP193095 | ASAP2 | 8853 | RNAseq | 0.2163 | 0.3779 | |
| SRP219564 | ASAP2 | 8853 | RNAseq | 0.0693 | 0.8876 | |
| TCGA | ASAP2 | 8853 | RNAseq | -0.0946 | 0.1432 |
Upregulated datasets: 1; Downregulated datasets: 0.
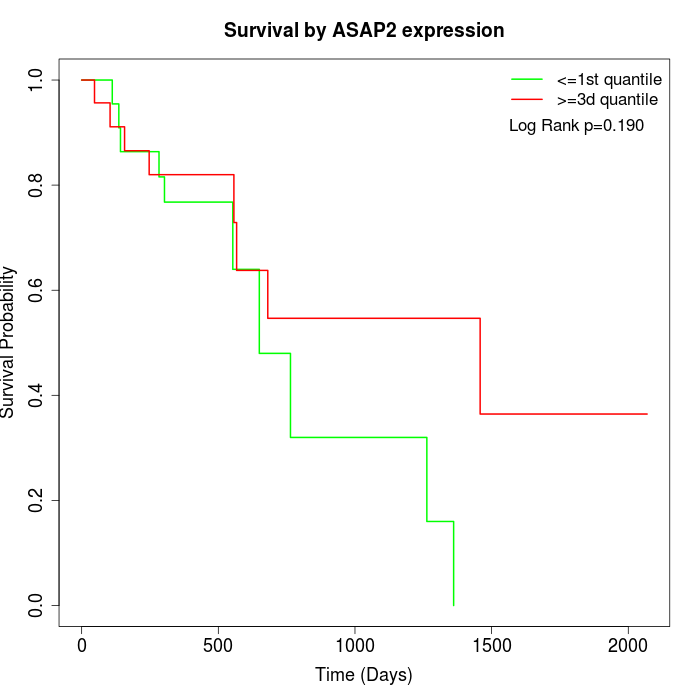
Survival by ASAP2 expression:
Note: Click image to view full size file.
Copy number change of ASAP2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ASAP2 | 8853 | 11 | 2 | 17 | |
| GSE20123 | ASAP2 | 8853 | 11 | 2 | 17 | |
| GSE43470 | ASAP2 | 8853 | 2 | 1 | 40 | |
| GSE46452 | ASAP2 | 8853 | 3 | 4 | 52 | |
| GSE47630 | ASAP2 | 8853 | 7 | 0 | 33 | |
| GSE54993 | ASAP2 | 8853 | 0 | 7 | 63 | |
| GSE54994 | ASAP2 | 8853 | 11 | 0 | 42 | |
| GSE60625 | ASAP2 | 8853 | 0 | 3 | 8 | |
| GSE74703 | ASAP2 | 8853 | 2 | 0 | 34 | |
| GSE74704 | ASAP2 | 8853 | 9 | 1 | 10 | |
| TCGA | ASAP2 | 8853 | 35 | 7 | 54 |
Total number of gains: 91; Total number of losses: 27; Total Number of normals: 370.
Somatic mutations of ASAP2:
Generating mutation plots.
Highly correlated genes for ASAP2:
Showing top 20/1045 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ASAP2 | TMTC1 | 0.761646 | 4 | 0 | 4 |
| ASAP2 | SNX18 | 0.758296 | 3 | 0 | 3 |
| ASAP2 | MZT1 | 0.755276 | 3 | 0 | 3 |
| ASAP2 | EXOSC6 | 0.754907 | 3 | 0 | 3 |
| ASAP2 | PDE7A | 0.749454 | 4 | 0 | 4 |
| ASAP2 | KBTBD6 | 0.745123 | 3 | 0 | 3 |
| ASAP2 | TRIB1 | 0.73817 | 3 | 0 | 3 |
| ASAP2 | TFAM | 0.725924 | 3 | 0 | 3 |
| ASAP2 | GZF1 | 0.724364 | 3 | 0 | 3 |
| ASAP2 | RILPL2 | 0.721407 | 3 | 0 | 3 |
| ASAP2 | GPATCH8 | 0.719045 | 3 | 0 | 3 |
| ASAP2 | RFX7 | 0.71695 | 3 | 0 | 3 |
| ASAP2 | CREB5 | 0.715192 | 3 | 0 | 3 |
| ASAP2 | SLCO3A1 | 0.713291 | 4 | 0 | 4 |
| ASAP2 | CPT1A | 0.713191 | 4 | 0 | 3 |
| ASAP2 | ELF4 | 0.71149 | 5 | 0 | 5 |
| ASAP2 | HSD17B1 | 0.71141 | 3 | 0 | 3 |
| ASAP2 | ACBD6 | 0.710772 | 4 | 0 | 4 |
| ASAP2 | DGUOK | 0.709117 | 8 | 0 | 7 |
| ASAP2 | CEP95 | 0.704769 | 3 | 0 | 3 |
For details and further investigation, click here