| Full name: transcription factor A, mitochondrial | Alias Symbol: | ||
| Type: protein-coding gene | Cytoband: 10q21.1 | ||
| Entrez ID: 7019 | HGNC ID: HGNC:11741 | Ensembl Gene: ENSG00000108064 | OMIM ID: 600438 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
Expression of TFAM:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TFAM | 7019 | 203177_x_at | 0.9332 | 0.1823 | |
| GSE20347 | TFAM | 7019 | 203177_x_at | 0.9129 | 0.0005 | |
| GSE23400 | TFAM | 7019 | 208541_x_at | 0.1743 | 0.0014 | |
| GSE26886 | TFAM | 7019 | 203177_x_at | 0.4168 | 0.1802 | |
| GSE29001 | TFAM | 7019 | 203177_x_at | 0.9027 | 0.0607 | |
| GSE38129 | TFAM | 7019 | 203177_x_at | 0.8993 | 0.0000 | |
| GSE45670 | TFAM | 7019 | 203177_x_at | 0.5048 | 0.1427 | |
| GSE53622 | TFAM | 7019 | 45918 | 0.3514 | 0.0000 | |
| GSE53624 | TFAM | 7019 | 45918 | 0.8531 | 0.0000 | |
| GSE63941 | TFAM | 7019 | 203177_x_at | 1.6267 | 0.0082 | |
| GSE77861 | TFAM | 7019 | 208541_x_at | 0.1522 | 0.3096 | |
| GSE97050 | TFAM | 7019 | A_33_P3389188 | 0.4348 | 0.2151 | |
| SRP007169 | TFAM | 7019 | RNAseq | 1.2267 | 0.0188 | |
| SRP008496 | TFAM | 7019 | RNAseq | 1.4699 | 0.0000 | |
| SRP064894 | TFAM | 7019 | RNAseq | 0.0145 | 0.9471 | |
| SRP133303 | TFAM | 7019 | RNAseq | 0.4961 | 0.0026 | |
| SRP159526 | TFAM | 7019 | RNAseq | 0.0372 | 0.9153 | |
| SRP193095 | TFAM | 7019 | RNAseq | 0.0427 | 0.6860 | |
| SRP219564 | TFAM | 7019 | RNAseq | 0.0770 | 0.8249 | |
| TCGA | TFAM | 7019 | RNAseq | 0.0664 | 0.2478 |
Upregulated datasets: 3; Downregulated datasets: 0.
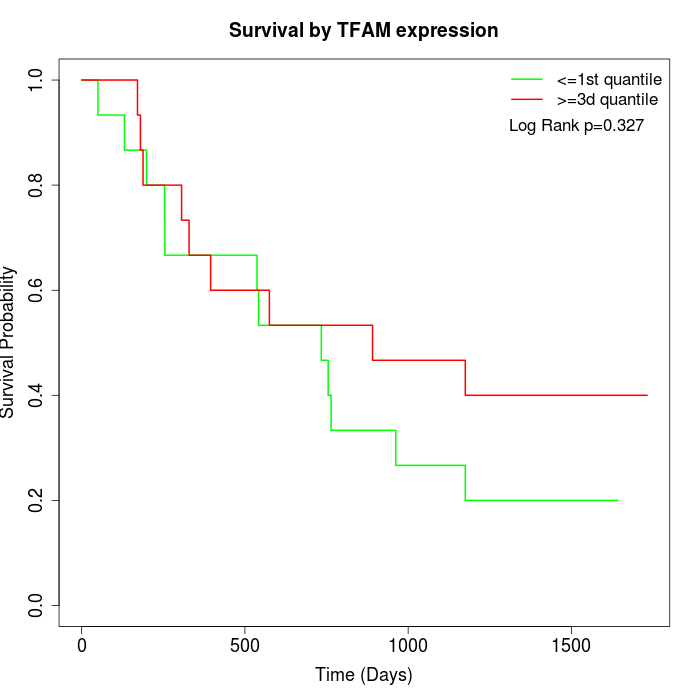
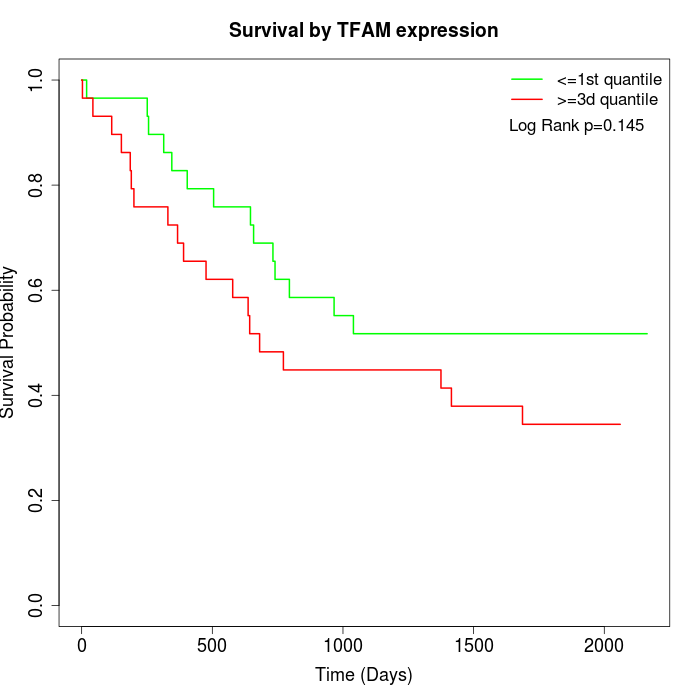
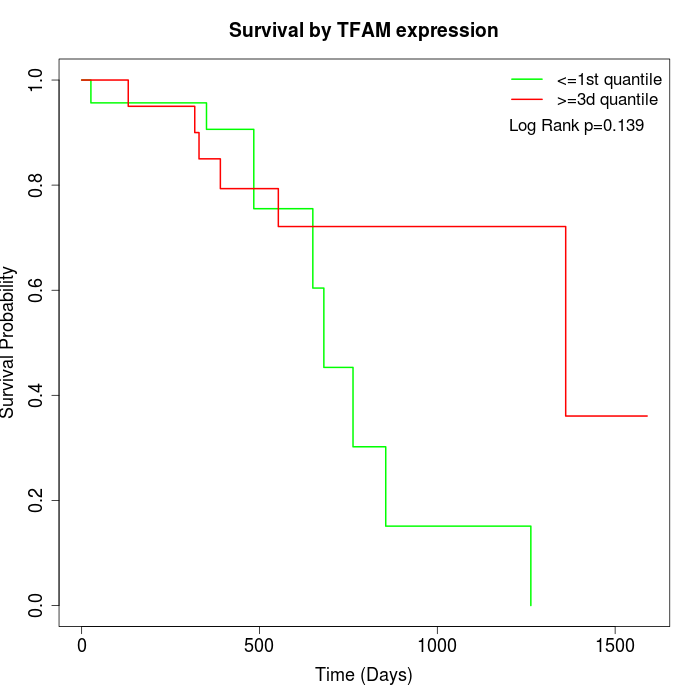
Survival by TFAM expression:
Note: Click image to view full size file.
Copy number change of TFAM:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TFAM | 7019 | 2 | 6 | 22 | |
| GSE20123 | TFAM | 7019 | 2 | 6 | 22 | |
| GSE43470 | TFAM | 7019 | 2 | 5 | 36 | |
| GSE46452 | TFAM | 7019 | 0 | 12 | 47 | |
| GSE47630 | TFAM | 7019 | 4 | 12 | 24 | |
| GSE54993 | TFAM | 7019 | 9 | 0 | 61 | |
| GSE54994 | TFAM | 7019 | 3 | 10 | 40 | |
| GSE60625 | TFAM | 7019 | 0 | 0 | 11 | |
| GSE74703 | TFAM | 7019 | 2 | 3 | 31 | |
| GSE74704 | TFAM | 7019 | 1 | 4 | 15 | |
| TCGA | TFAM | 7019 | 10 | 23 | 63 |
Total number of gains: 35; Total number of losses: 81; Total Number of normals: 372.
Somatic mutations of TFAM:
Generating mutation plots.
Highly correlated genes for TFAM:
Showing top 20/1371 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TFAM | GPR107 | 0.750213 | 3 | 0 | 3 |
| TFAM | FHL3 | 0.745218 | 3 | 0 | 3 |
| TFAM | CCDC124 | 0.727319 | 3 | 0 | 3 |
| TFAM | ASAP2 | 0.725924 | 3 | 0 | 3 |
| TFAM | ZNF707 | 0.721739 | 3 | 0 | 3 |
| TFAM | FCHO1 | 0.71981 | 3 | 0 | 3 |
| TFAM | FUBP3 | 0.717165 | 3 | 0 | 3 |
| TFAM | TCP1 | 0.710525 | 3 | 0 | 3 |
| TFAM | HPS3 | 0.707045 | 3 | 0 | 3 |
| TFAM | PTCD1 | 0.704177 | 3 | 0 | 3 |
| TFAM | TOMM20 | 0.701166 | 6 | 0 | 6 |
| TFAM | MAD2L2 | 0.692569 | 6 | 0 | 5 |
| TFAM | MMS22L | 0.692123 | 3 | 0 | 3 |
| TFAM | SLC35B2 | 0.69145 | 3 | 0 | 3 |
| TFAM | NF1 | 0.69083 | 3 | 0 | 3 |
| TFAM | PSMD1 | 0.687648 | 3 | 0 | 3 |
| TFAM | PRKRA | 0.68703 | 3 | 0 | 3 |
| TFAM | PRDX4 | 0.686504 | 3 | 0 | 3 |
| TFAM | IMPACT | 0.68636 | 3 | 0 | 3 |
| TFAM | ATP2C2 | 0.684284 | 3 | 0 | 3 |
For details and further investigation, click here