| Full name: ATPase H+ transporting V1 subunit D | Alias Symbol: VATD|VMA8 | ||
| Type: protein-coding gene | Cytoband: 14q23.3 | ||
| Entrez ID: 51382 | HGNC ID: HGNC:13527 | Ensembl Gene: ENSG00000100554 | OMIM ID: 609398 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ATP6V1D involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04150 | mTOR signaling pathway | |
| hsa05110 | Vibrio cholerae infection | |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection |
Expression of ATP6V1D:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATP6V1D | 51382 | 208898_at | -0.9760 | 0.1212 | |
| GSE20347 | ATP6V1D | 51382 | 208898_at | -1.3890 | 0.0000 | |
| GSE23400 | ATP6V1D | 51382 | 208898_at | -0.9269 | 0.0000 | |
| GSE26886 | ATP6V1D | 51382 | 208898_at | -1.7629 | 0.0000 | |
| GSE29001 | ATP6V1D | 51382 | 208898_at | -0.8487 | 0.0028 | |
| GSE38129 | ATP6V1D | 51382 | 208898_at | -0.9999 | 0.0000 | |
| GSE45670 | ATP6V1D | 51382 | 208898_at | -0.2928 | 0.1659 | |
| GSE53622 | ATP6V1D | 51382 | 101470 | -0.6984 | 0.0000 | |
| GSE53624 | ATP6V1D | 51382 | 101470 | -0.6641 | 0.0000 | |
| GSE63941 | ATP6V1D | 51382 | 208899_x_at | -0.1373 | 0.7419 | |
| GSE77861 | ATP6V1D | 51382 | 208898_at | -1.3144 | 0.0032 | |
| GSE97050 | ATP6V1D | 51382 | A_33_P3387463 | -0.4209 | 0.2141 | |
| SRP007169 | ATP6V1D | 51382 | RNAseq | -2.6105 | 0.0000 | |
| SRP008496 | ATP6V1D | 51382 | RNAseq | -2.3547 | 0.0000 | |
| SRP064894 | ATP6V1D | 51382 | RNAseq | -1.4664 | 0.0000 | |
| SRP133303 | ATP6V1D | 51382 | RNAseq | -0.9526 | 0.0000 | |
| SRP159526 | ATP6V1D | 51382 | RNAseq | -1.7668 | 0.0000 | |
| SRP193095 | ATP6V1D | 51382 | RNAseq | -1.6260 | 0.0000 | |
| SRP219564 | ATP6V1D | 51382 | RNAseq | -1.4570 | 0.0025 | |
| TCGA | ATP6V1D | 51382 | RNAseq | -0.0286 | 0.6241 |
Upregulated datasets: 0; Downregulated datasets: 9.
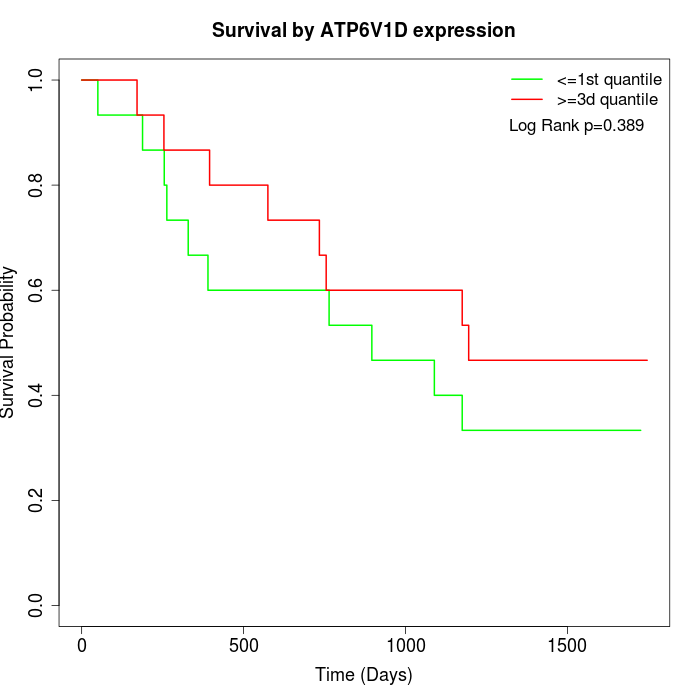
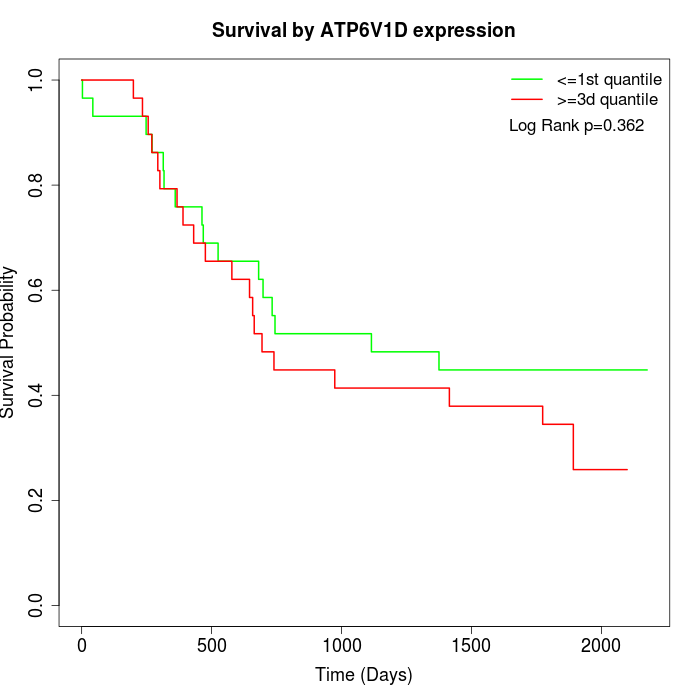
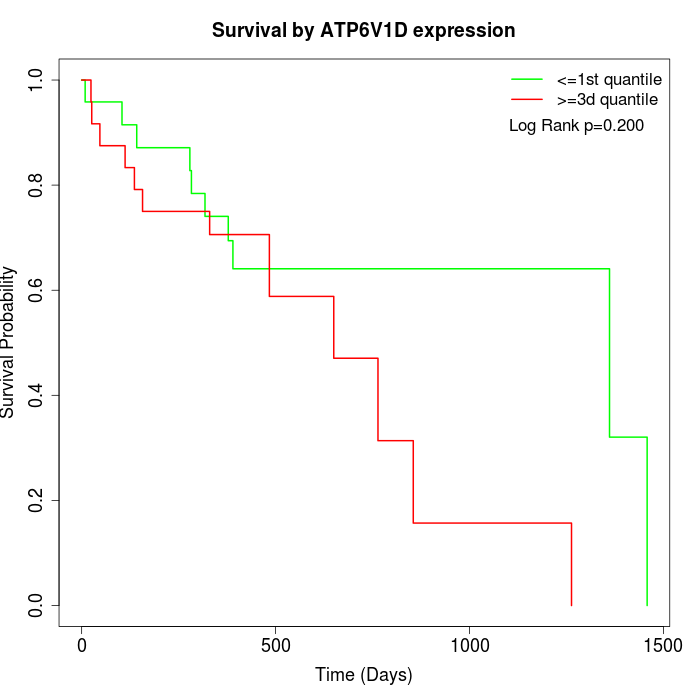
Survival by ATP6V1D expression:
Note: Click image to view full size file.
Copy number change of ATP6V1D:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATP6V1D | 51382 | 7 | 3 | 20 | |
| GSE20123 | ATP6V1D | 51382 | 7 | 3 | 20 | |
| GSE43470 | ATP6V1D | 51382 | 8 | 2 | 33 | |
| GSE46452 | ATP6V1D | 51382 | 16 | 3 | 40 | |
| GSE47630 | ATP6V1D | 51382 | 11 | 9 | 20 | |
| GSE54993 | ATP6V1D | 51382 | 3 | 9 | 58 | |
| GSE54994 | ATP6V1D | 51382 | 18 | 5 | 30 | |
| GSE60625 | ATP6V1D | 51382 | 0 | 2 | 9 | |
| GSE74703 | ATP6V1D | 51382 | 7 | 2 | 27 | |
| GSE74704 | ATP6V1D | 51382 | 3 | 2 | 15 | |
| TCGA | ATP6V1D | 51382 | 35 | 14 | 47 |
Total number of gains: 115; Total number of losses: 54; Total Number of normals: 319.
Somatic mutations of ATP6V1D:
Generating mutation plots.
Highly correlated genes for ATP6V1D:
Showing top 20/1598 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATP6V1D | EPG5 | 0.811784 | 4 | 0 | 4 |
| ATP6V1D | CES2 | 0.805835 | 8 | 0 | 8 |
| ATP6V1D | ZNF554 | 0.797251 | 3 | 0 | 3 |
| ATP6V1D | NPEPPS | 0.795643 | 10 | 0 | 10 |
| ATP6V1D | PDCD6IP | 0.789146 | 11 | 0 | 11 |
| ATP6V1D | C18orf25 | 0.782127 | 10 | 0 | 10 |
| ATP6V1D | TRIP10 | 0.781776 | 10 | 0 | 10 |
| ATP6V1D | ITCH | 0.780454 | 10 | 0 | 9 |
| ATP6V1D | VSIG10L | 0.778503 | 5 | 0 | 5 |
| ATP6V1D | SFTA2 | 0.77827 | 4 | 0 | 4 |
| ATP6V1D | CTTNBP2NL | 0.769627 | 5 | 0 | 4 |
| ATP6V1D | WDFY1 | 0.768057 | 3 | 0 | 3 |
| ATP6V1D | EXOC6B | 0.767026 | 3 | 0 | 3 |
| ATP6V1D | CWH43 | 0.764502 | 10 | 0 | 8 |
| ATP6V1D | NAGK | 0.764231 | 10 | 0 | 10 |
| ATP6V1D | ECM1 | 0.763038 | 10 | 0 | 9 |
| ATP6V1D | SNX24 | 0.761897 | 9 | 0 | 9 |
| ATP6V1D | ACAA1 | 0.76152 | 10 | 0 | 9 |
| ATP6V1D | CAST | 0.761411 | 11 | 0 | 10 |
| ATP6V1D | PRSS2 | 0.761044 | 8 | 0 | 8 |
For details and further investigation, click here