| Full name: itchy E3 ubiquitin protein ligase | Alias Symbol: AIP4 | ||
| Type: protein-coding gene | Cytoband: 20q11.22 | ||
| Entrez ID: 83737 | HGNC ID: HGNC:13890 | Ensembl Gene: ENSG00000078747 | OMIM ID: 606409 |
| Drug and gene relationship at DGIdb | |||
ITCH involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04668 | TNF signaling pathway | |
| hsa04932 | Non-alcoholic fatty liver disease (NAFLD) |
Expression of ITCH:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ITCH | 83737 | 209744_x_at | -0.6667 | 0.4463 | |
| GSE20347 | ITCH | 83737 | 209744_x_at | -1.5446 | 0.0000 | |
| GSE23400 | ITCH | 83737 | 209744_x_at | -0.9735 | 0.0000 | |
| GSE26886 | ITCH | 83737 | 209744_x_at | -2.0718 | 0.0000 | |
| GSE29001 | ITCH | 83737 | 209744_x_at | -1.3698 | 0.0061 | |
| GSE38129 | ITCH | 83737 | 209744_x_at | -0.9974 | 0.0038 | |
| GSE45670 | ITCH | 83737 | 209744_x_at | -0.7299 | 0.0108 | |
| GSE53622 | ITCH | 83737 | 16407 | -0.4970 | 0.0000 | |
| GSE53624 | ITCH | 83737 | 16407 | -0.7257 | 0.0000 | |
| GSE63941 | ITCH | 83737 | 209744_x_at | 0.4539 | 0.4097 | |
| GSE77861 | ITCH | 83737 | 209744_x_at | -1.1668 | 0.0287 | |
| GSE97050 | ITCH | 83737 | A_24_P139191 | -0.1036 | 0.6436 | |
| SRP007169 | ITCH | 83737 | RNAseq | -0.7191 | 0.0676 | |
| SRP008496 | ITCH | 83737 | RNAseq | -0.5046 | 0.0151 | |
| SRP064894 | ITCH | 83737 | RNAseq | -1.2156 | 0.0000 | |
| SRP133303 | ITCH | 83737 | RNAseq | -0.3276 | 0.0053 | |
| SRP159526 | ITCH | 83737 | RNAseq | -1.1974 | 0.0000 | |
| SRP193095 | ITCH | 83737 | RNAseq | -0.8350 | 0.0000 | |
| SRP219564 | ITCH | 83737 | RNAseq | -1.0169 | 0.0282 | |
| TCGA | ITCH | 83737 | RNAseq | -0.0441 | 0.3304 |
Upregulated datasets: 0; Downregulated datasets: 7.
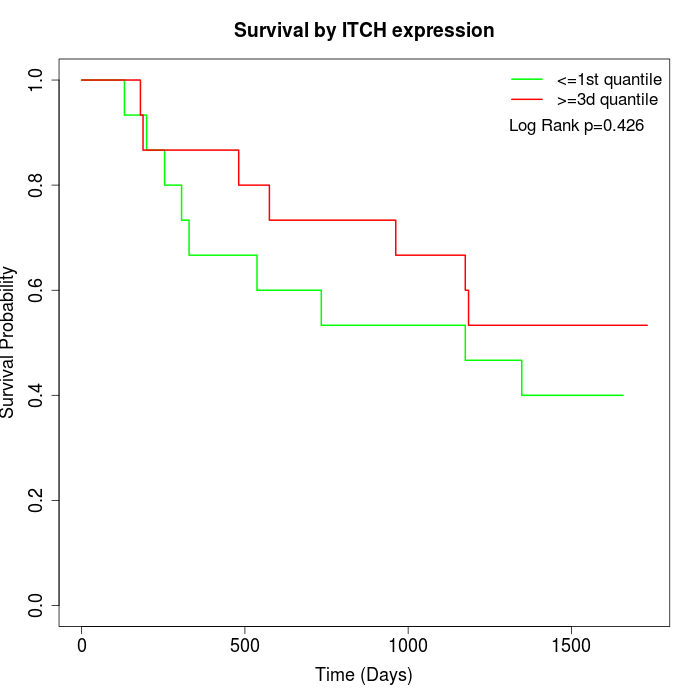
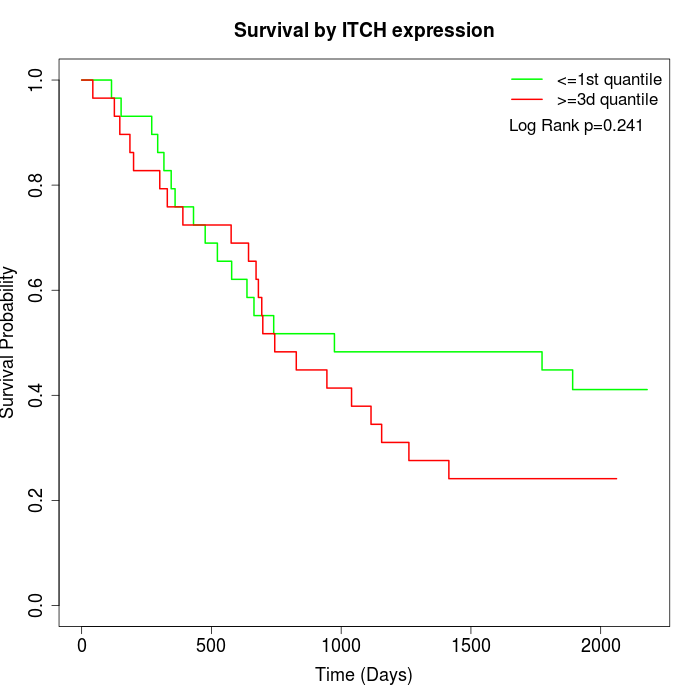
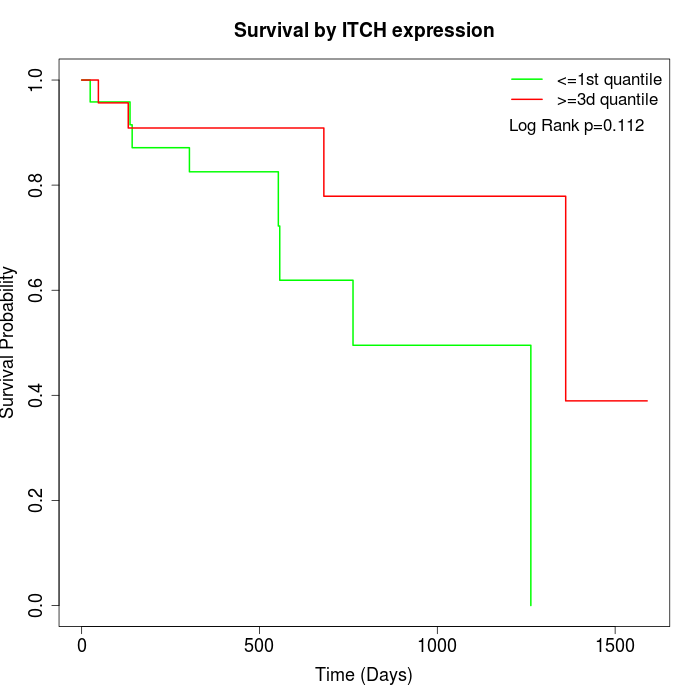
Survival by ITCH expression:
Note: Click image to view full size file.
Copy number change of ITCH:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ITCH | 83737 | 16 | 0 | 14 | |
| GSE20123 | ITCH | 83737 | 16 | 0 | 14 | |
| GSE43470 | ITCH | 83737 | 12 | 0 | 31 | |
| GSE46452 | ITCH | 83737 | 29 | 0 | 30 | |
| GSE47630 | ITCH | 83737 | 24 | 1 | 15 | |
| GSE54993 | ITCH | 83737 | 0 | 18 | 52 | |
| GSE54994 | ITCH | 83737 | 25 | 1 | 27 | |
| GSE60625 | ITCH | 83737 | 0 | 0 | 11 | |
| GSE74703 | ITCH | 83737 | 11 | 0 | 25 | |
| GSE74704 | ITCH | 83737 | 11 | 0 | 9 | |
| TCGA | ITCH | 83737 | 51 | 1 | 44 |
Total number of gains: 195; Total number of losses: 21; Total Number of normals: 272.
Somatic mutations of ITCH:
Generating mutation plots.
Highly correlated genes for ITCH:
Showing top 20/1567 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ITCH | SPNS2 | 0.805186 | 4 | 0 | 4 |
| ITCH | SFTA2 | 0.79788 | 4 | 0 | 4 |
| ITCH | SH3GLB1 | 0.796734 | 10 | 0 | 10 |
| ITCH | ELMO2 | 0.79624 | 10 | 0 | 10 |
| ITCH | LEPROT | 0.79354 | 3 | 0 | 3 |
| ITCH | ANKRD37 | 0.788848 | 5 | 0 | 5 |
| ITCH | ZFC3H1 | 0.783616 | 3 | 0 | 3 |
| ITCH | C18orf25 | 0.782392 | 11 | 0 | 11 |
| ITCH | ABHD5 | 0.78086 | 10 | 0 | 10 |
| ITCH | ATP6V1D | 0.780454 | 10 | 0 | 9 |
| ITCH | CTTNBP2NL | 0.765031 | 7 | 0 | 7 |
| ITCH | STRN | 0.764946 | 7 | 0 | 7 |
| ITCH | CPEB4 | 0.761695 | 6 | 0 | 6 |
| ITCH | PDCD6IP | 0.760717 | 10 | 0 | 9 |
| ITCH | ATP6V1C2 | 0.755944 | 4 | 0 | 4 |
| ITCH | CDK7 | 0.754665 | 10 | 0 | 10 |
| ITCH | ZNF426 | 0.749688 | 10 | 0 | 10 |
| ITCH | WWC1 | 0.749248 | 9 | 0 | 9 |
| ITCH | ZMYM2 | 0.748894 | 4 | 0 | 4 |
| ITCH | CCNYL1 | 0.748281 | 5 | 0 | 4 |
For details and further investigation, click here