| Full name: ATPase H+ transporting V1 subunit E2 | Alias Symbol: MGC9341|VMA4|ATP6E1 | ||
| Type: protein-coding gene | Cytoband: 2p21 | ||
| Entrez ID: 90423 | HGNC ID: HGNC:18125 | Ensembl Gene: ENSG00000250565 | OMIM ID: 617385 |
| Drug and gene relationship at DGIdb | |||
ATP6V1E2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04150 | mTOR signaling pathway | |
| hsa05110 | Vibrio cholerae infection | |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection |
Expression of ATP6V1E2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATP6V1E2 | 90423 | 1552286_at | 0.7651 | 0.3826 | |
| GSE26886 | ATP6V1E2 | 90423 | 1552286_at | 1.0674 | 0.0006 | |
| GSE45670 | ATP6V1E2 | 90423 | 1552286_at | 0.8697 | 0.0124 | |
| GSE53622 | ATP6V1E2 | 90423 | 96318 | 1.1401 | 0.0000 | |
| GSE53624 | ATP6V1E2 | 90423 | 96318 | 1.3200 | 0.0000 | |
| GSE63941 | ATP6V1E2 | 90423 | 1552286_at | 0.8388 | 0.1102 | |
| GSE77861 | ATP6V1E2 | 90423 | 1552286_at | 0.3610 | 0.0697 | |
| GSE97050 | ATP6V1E2 | A_21_P0014708 | 0.0243 | 0.9295 | ||
| SRP007169 | ATP6V1E2 | 90423 | RNAseq | 0.1787 | 0.7716 | |
| SRP008496 | ATP6V1E2 | 90423 | RNAseq | 1.5150 | 0.0053 | |
| SRP064894 | ATP6V1E2 | 90423 | RNAseq | 1.0723 | 0.0000 | |
| SRP133303 | ATP6V1E2 | 90423 | RNAseq | 0.8670 | 0.0196 | |
| SRP159526 | ATP6V1E2 | 90423 | RNAseq | 1.3071 | 0.0006 | |
| SRP193095 | ATP6V1E2 | 90423 | RNAseq | 1.1236 | 0.0000 | |
| SRP219564 | ATP6V1E2 | 90423 | RNAseq | 1.3343 | 0.0348 | |
| TCGA | ATP6V1E2 | 90423 | RNAseq | 0.5214 | 0.0000 |
Upregulated datasets: 8; Downregulated datasets: 0.
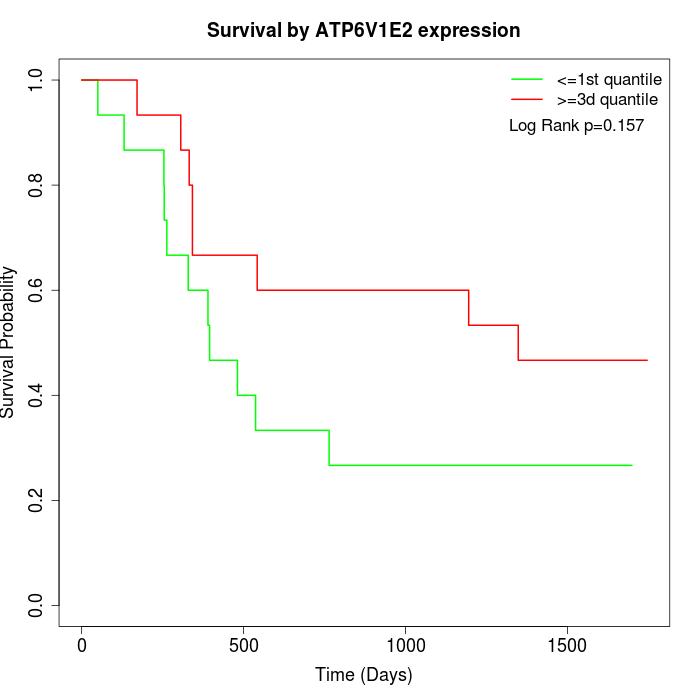
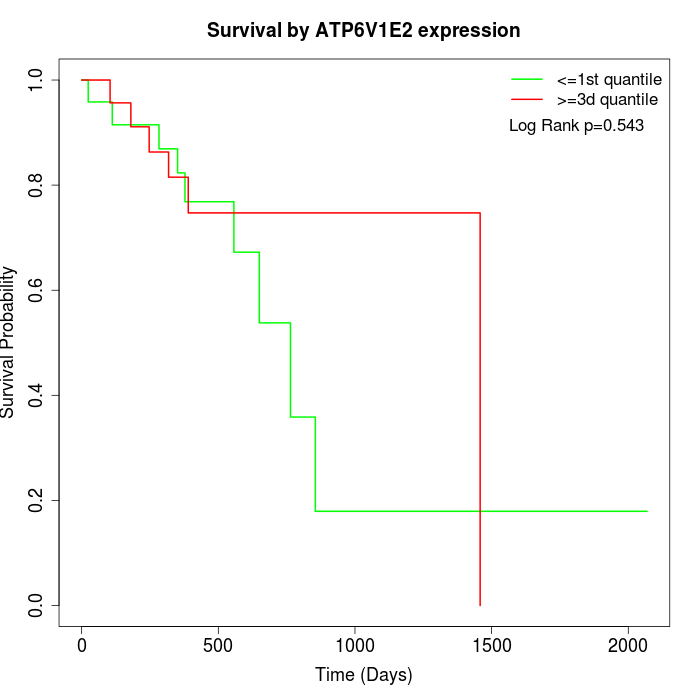
Survival by ATP6V1E2 expression:
Note: Click image to view full size file.
Copy number change of ATP6V1E2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATP6V1E2 | 90423 | 10 | 1 | 19 | |
| GSE20123 | ATP6V1E2 | 90423 | 9 | 1 | 20 | |
| GSE43470 | ATP6V1E2 | 90423 | 5 | 0 | 38 | |
| GSE46452 | ATP6V1E2 | 90423 | 1 | 4 | 54 | |
| GSE47630 | ATP6V1E2 | 90423 | 7 | 0 | 33 | |
| GSE54993 | ATP6V1E2 | 90423 | 0 | 5 | 65 | |
| GSE54994 | ATP6V1E2 | 90423 | 10 | 0 | 43 | |
| GSE60625 | ATP6V1E2 | 90423 | 0 | 3 | 8 | |
| GSE74703 | ATP6V1E2 | 90423 | 5 | 0 | 31 | |
| GSE74704 | ATP6V1E2 | 90423 | 9 | 1 | 10 | |
| TCGA | ATP6V1E2 | 90423 | 35 | 2 | 59 |
Total number of gains: 91; Total number of losses: 17; Total Number of normals: 380.
Somatic mutations of ATP6V1E2:
Generating mutation plots.
Highly correlated genes for ATP6V1E2:
Showing top 20/1332 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATP6V1E2 | STON1 | 0.771259 | 3 | 0 | 3 |
| ATP6V1E2 | NOS1AP | 0.734329 | 3 | 0 | 3 |
| ATP6V1E2 | HTRA2 | 0.712601 | 6 | 0 | 6 |
| ATP6V1E2 | RILPL2 | 0.704207 | 4 | 0 | 3 |
| ATP6V1E2 | FADS1 | 0.70386 | 3 | 0 | 3 |
| ATP6V1E2 | PYCR2 | 0.701415 | 3 | 0 | 3 |
| ATP6V1E2 | CPT1A | 0.701258 | 4 | 0 | 3 |
| ATP6V1E2 | GALNT18 | 0.699942 | 6 | 0 | 6 |
| ATP6V1E2 | LRCH3 | 0.696336 | 3 | 0 | 3 |
| ATP6V1E2 | MB21D2 | 0.696222 | 6 | 0 | 5 |
| ATP6V1E2 | VPS13C | 0.693422 | 3 | 0 | 3 |
| ATP6V1E2 | CAD | 0.692432 | 8 | 0 | 8 |
| ATP6V1E2 | DGKH | 0.692155 | 4 | 0 | 3 |
| ATP6V1E2 | HAUS1 | 0.691894 | 4 | 0 | 4 |
| ATP6V1E2 | CHD1 | 0.691594 | 3 | 0 | 3 |
| ATP6V1E2 | SRXN1 | 0.690564 | 4 | 0 | 4 |
| ATP6V1E2 | RAB34 | 0.690403 | 3 | 0 | 3 |
| ATP6V1E2 | LRRC58 | 0.689467 | 4 | 0 | 3 |
| ATP6V1E2 | SDR39U1 | 0.689191 | 3 | 0 | 3 |
| ATP6V1E2 | METTL23 | 0.686352 | 6 | 0 | 6 |
For details and further investigation, click here