| Full name: ATPase H+ transporting V1 subunit F | Alias Symbol: ATP6S14|VATF|Vma7 | ||
| Type: protein-coding gene | Cytoband: 7q32.1 | ||
| Entrez ID: 9296 | HGNC ID: HGNC:16832 | Ensembl Gene: ENSG00000128524 | OMIM ID: 607160 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
ATP6V1F involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04150 | mTOR signaling pathway | |
| hsa05110 | Vibrio cholerae infection | |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection |
Expression of ATP6V1F:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATP6V1F | 9296 | 201527_at | -0.1559 | 0.6508 | |
| GSE20347 | ATP6V1F | 9296 | 201527_at | -0.4518 | 0.0001 | |
| GSE23400 | ATP6V1F | 9296 | 201527_at | -0.1813 | 0.0077 | |
| GSE26886 | ATP6V1F | 9296 | 201527_at | -0.6282 | 0.0023 | |
| GSE29001 | ATP6V1F | 9296 | 201527_at | -0.3459 | 0.2335 | |
| GSE38129 | ATP6V1F | 9296 | 201527_at | -0.2034 | 0.2616 | |
| GSE45670 | ATP6V1F | 9296 | 201527_at | -0.2501 | 0.0969 | |
| GSE53622 | ATP6V1F | 9296 | 37208 | -0.4152 | 0.0000 | |
| GSE53624 | ATP6V1F | 9296 | 37208 | -0.4321 | 0.0000 | |
| GSE63941 | ATP6V1F | 9296 | 201527_at | -0.3447 | 0.4015 | |
| GSE77861 | ATP6V1F | 9296 | 201527_at | -0.4311 | 0.0244 | |
| GSE97050 | ATP6V1F | 9296 | A_23_P93623 | -0.0863 | 0.7206 | |
| SRP007169 | ATP6V1F | 9296 | RNAseq | -1.1241 | 0.0011 | |
| SRP008496 | ATP6V1F | 9296 | RNAseq | -1.1092 | 0.0000 | |
| SRP064894 | ATP6V1F | 9296 | RNAseq | -0.1600 | 0.5588 | |
| SRP133303 | ATP6V1F | 9296 | RNAseq | -0.3775 | 0.0183 | |
| SRP159526 | ATP6V1F | 9296 | RNAseq | -0.5991 | 0.0758 | |
| SRP193095 | ATP6V1F | 9296 | RNAseq | -0.8852 | 0.0000 | |
| SRP219564 | ATP6V1F | 9296 | RNAseq | -0.8254 | 0.0119 | |
| TCGA | ATP6V1F | 9296 | RNAseq | 0.1561 | 0.0123 |
Upregulated datasets: 0; Downregulated datasets: 2.
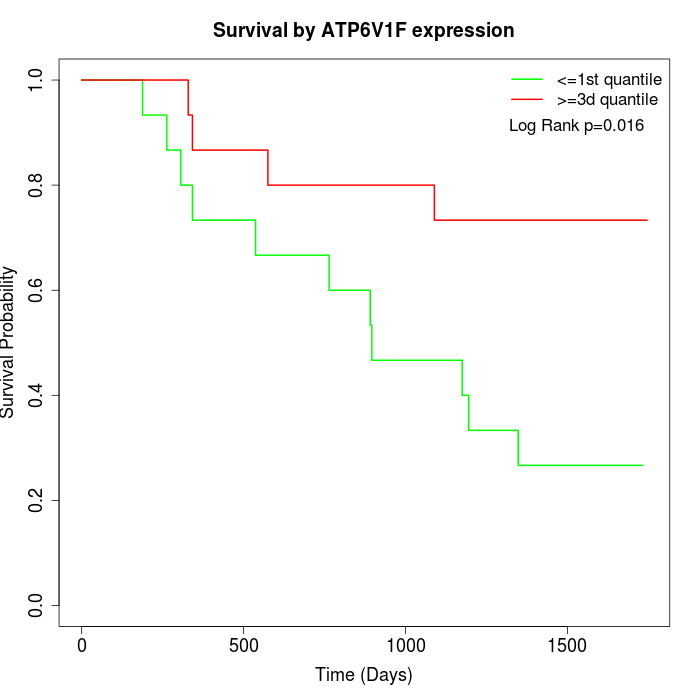
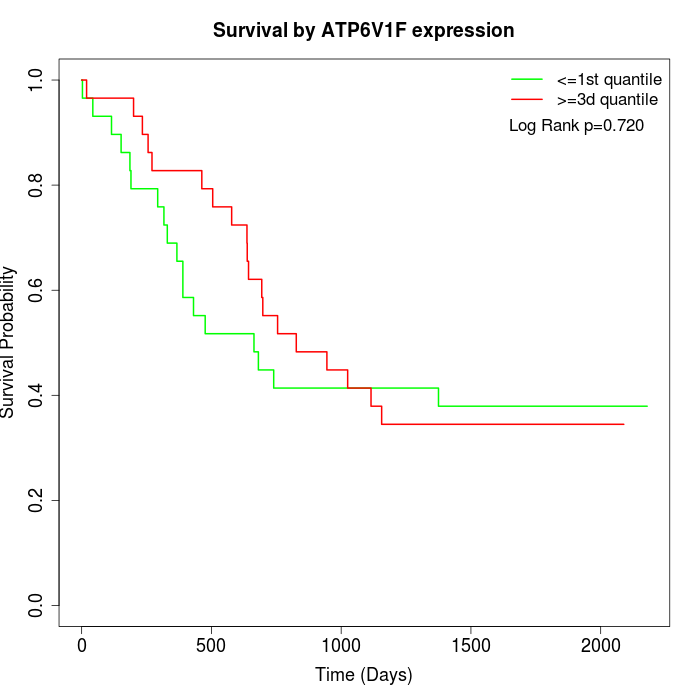
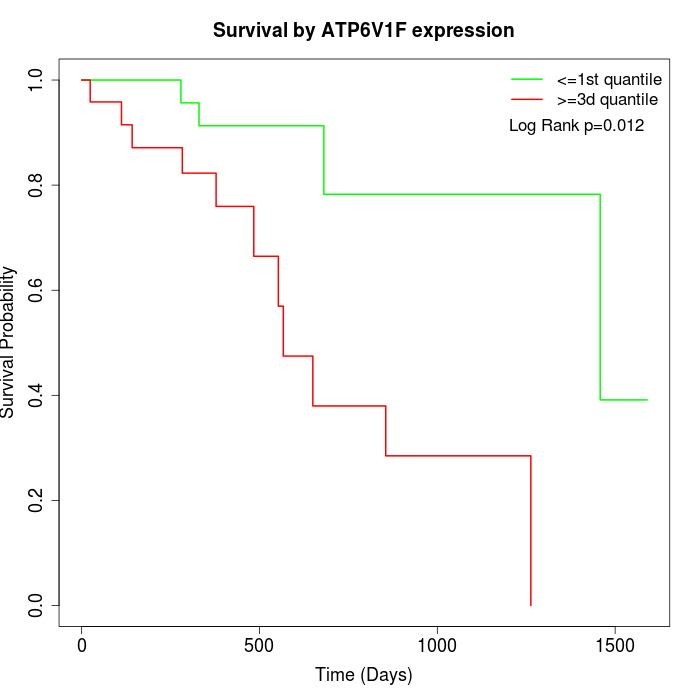
Survival by ATP6V1F expression:
Note: Click image to view full size file.
Copy number change of ATP6V1F:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATP6V1F | 9296 | 8 | 1 | 21 | |
| GSE20123 | ATP6V1F | 9296 | 8 | 1 | 21 | |
| GSE43470 | ATP6V1F | 9296 | 2 | 3 | 38 | |
| GSE46452 | ATP6V1F | 9296 | 7 | 2 | 50 | |
| GSE47630 | ATP6V1F | 9296 | 7 | 4 | 29 | |
| GSE54993 | ATP6V1F | 9296 | 3 | 5 | 62 | |
| GSE54994 | ATP6V1F | 9296 | 8 | 7 | 38 | |
| GSE60625 | ATP6V1F | 9296 | 0 | 0 | 11 | |
| GSE74703 | ATP6V1F | 9296 | 2 | 3 | 31 | |
| GSE74704 | ATP6V1F | 9296 | 4 | 1 | 15 | |
| TCGA | ATP6V1F | 9296 | 28 | 22 | 46 |
Total number of gains: 77; Total number of losses: 49; Total Number of normals: 362.
Somatic mutations of ATP6V1F:
Generating mutation plots.
Highly correlated genes for ATP6V1F:
Showing top 20/712 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATP6V1F | NCCRP1 | 0.745935 | 3 | 0 | 3 |
| ATP6V1F | SDCBP2 | 0.720097 | 3 | 0 | 3 |
| ATP6V1F | TBC1D10A | 0.719164 | 3 | 0 | 3 |
| ATP6V1F | HES5 | 0.712883 | 3 | 0 | 3 |
| ATP6V1F | TECPR1 | 0.711744 | 4 | 0 | 3 |
| ATP6V1F | ZNF385A | 0.694594 | 3 | 0 | 3 |
| ATP6V1F | ARF5 | 0.691814 | 12 | 0 | 10 |
| ATP6V1F | TMPRSS11A | 0.689474 | 3 | 0 | 3 |
| ATP6V1F | SPAG17 | 0.680828 | 3 | 0 | 3 |
| ATP6V1F | UPK3B | 0.680518 | 3 | 0 | 3 |
| ATP6V1F | BARX2 | 0.670218 | 8 | 0 | 8 |
| ATP6V1F | RNF222 | 0.669874 | 3 | 0 | 3 |
| ATP6V1F | METTL25 | 0.668372 | 3 | 0 | 3 |
| ATP6V1F | TPRN | 0.667053 | 6 | 0 | 4 |
| ATP6V1F | TMED3 | 0.664647 | 10 | 0 | 10 |
| ATP6V1F | TSTD1 | 0.664563 | 5 | 0 | 4 |
| ATP6V1F | TRIM29 | 0.663276 | 11 | 0 | 11 |
| ATP6V1F | CTNNBIP1 | 0.663036 | 9 | 0 | 8 |
| ATP6V1F | TMEM141 | 0.659107 | 5 | 0 | 5 |
| ATP6V1F | BTBD9 | 0.658786 | 3 | 0 | 3 |
For details and further investigation, click here