| Full name: ATPase H+ transporting V1 subunit G2 | Alias Symbol: Vma10|NG38|Em:AC004181.3 | ||
| Type: protein-coding gene | Cytoband: 6p21.33 | ||
| Entrez ID: 534 | HGNC ID: HGNC:862 | Ensembl Gene: ENSG00000213760 | OMIM ID: 606853 |
| Drug and gene relationship at DGIdb | |||
ATP6V1G2 involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04150 | mTOR signaling pathway | |
| hsa05110 | Vibrio cholerae infection | |
| hsa05120 | Epithelial cell signaling in Helicobacter pylori infection |
Expression of ATP6V1G2:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | ATP6V1G2 | 534 | 214762_at | -0.2518 | 0.5691 | |
| GSE20347 | ATP6V1G2 | 534 | 214762_at | -0.0343 | 0.6684 | |
| GSE23400 | ATP6V1G2 | 534 | 214762_at | -0.2121 | 0.0000 | |
| GSE26886 | ATP6V1G2 | 534 | 214762_at | -0.2634 | 0.0347 | |
| GSE29001 | ATP6V1G2 | 534 | 214762_at | -0.0344 | 0.8626 | |
| GSE38129 | ATP6V1G2 | 534 | 214762_at | -0.2316 | 0.0121 | |
| GSE45670 | ATP6V1G2 | 534 | 214762_at | -0.3249 | 0.0209 | |
| GSE53622 | ATP6V1G2 | 534 | 67067 | -0.5756 | 0.0003 | |
| GSE53624 | ATP6V1G2 | 534 | 67067 | -0.4117 | 0.0028 | |
| GSE63941 | ATP6V1G2 | 534 | 214762_at | -0.7215 | 0.0005 | |
| GSE77861 | ATP6V1G2 | 534 | 214762_at | -0.1456 | 0.1340 | |
| GSE97050 | ATP6V1G2 | 534 | A_23_P500410 | -0.3529 | 0.2365 | |
| SRP159526 | ATP6V1G2 | 534 | RNAseq | -0.2172 | 0.7886 | |
| TCGA | ATP6V1G2 | 534 | RNAseq | -1.3150 | 0.0000 |
Upregulated datasets: 0; Downregulated datasets: 1.
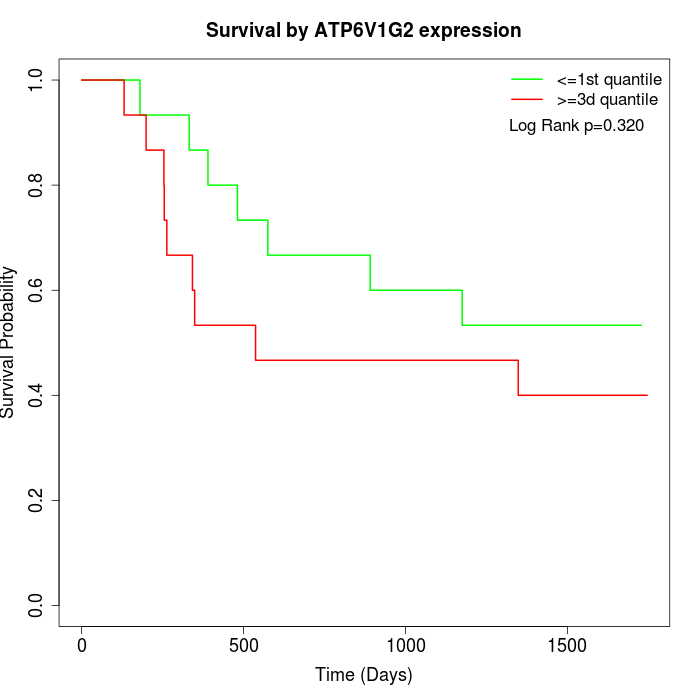
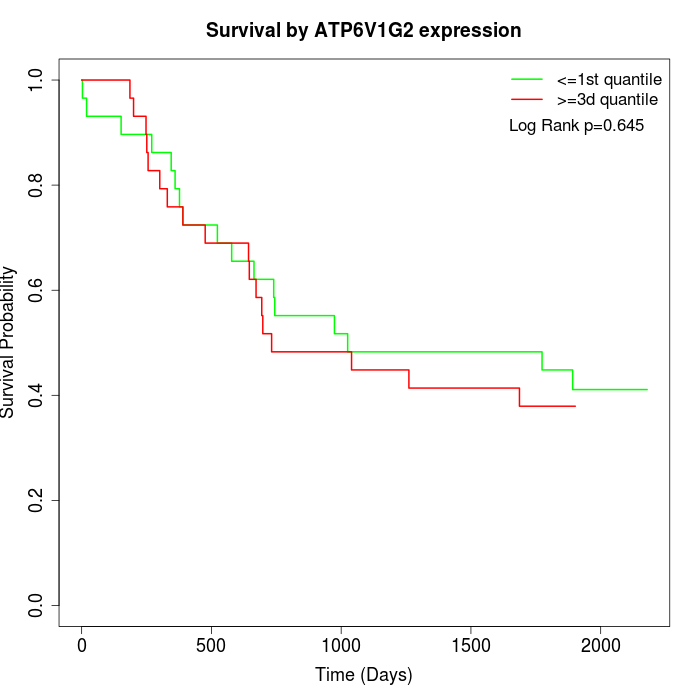
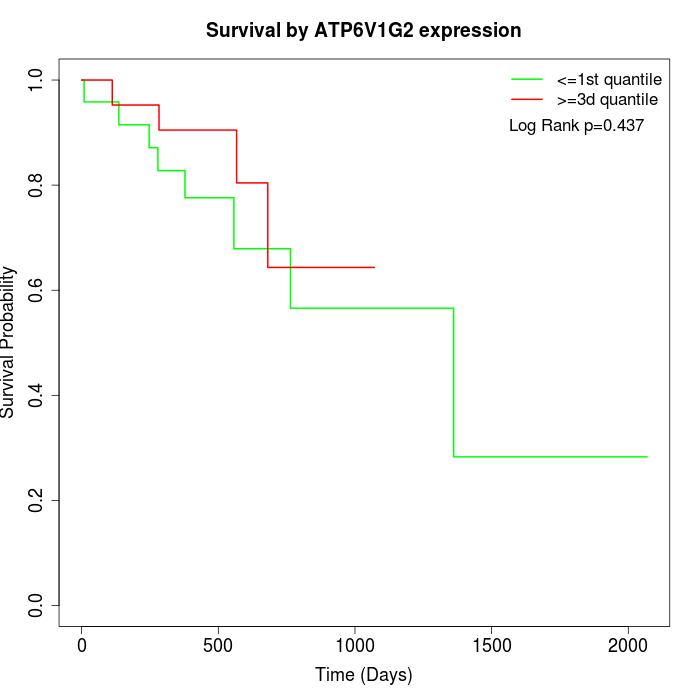
Survival by ATP6V1G2 expression:
Note: Click image to view full size file.
Copy number change of ATP6V1G2:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | ATP6V1G2 | 534 | 5 | 1 | 24 | |
| GSE20123 | ATP6V1G2 | 534 | 5 | 1 | 24 | |
| GSE43470 | ATP6V1G2 | 534 | 6 | 3 | 34 | |
| GSE46452 | ATP6V1G2 | 534 | 1 | 10 | 48 | |
| GSE47630 | ATP6V1G2 | 534 | 7 | 5 | 28 | |
| GSE54993 | ATP6V1G2 | 534 | 2 | 1 | 67 | |
| GSE54994 | ATP6V1G2 | 534 | 11 | 4 | 38 | |
| GSE60625 | ATP6V1G2 | 534 | 0 | 1 | 10 | |
| GSE74703 | ATP6V1G2 | 534 | 6 | 1 | 29 | |
| GSE74704 | ATP6V1G2 | 534 | 2 | 0 | 18 | |
| TCGA | ATP6V1G2 | 534 | 16 | 16 | 64 |
Total number of gains: 61; Total number of losses: 43; Total Number of normals: 384.
Somatic mutations of ATP6V1G2:
Generating mutation plots.
Highly correlated genes for ATP6V1G2:
Showing top 20/756 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| ATP6V1G2 | CNTF | 0.762315 | 3 | 0 | 3 |
| ATP6V1G2 | NDUFA11 | 0.757649 | 3 | 0 | 3 |
| ATP6V1G2 | KLHL18 | 0.748742 | 3 | 0 | 3 |
| ATP6V1G2 | IFNA5 | 0.739633 | 3 | 0 | 3 |
| ATP6V1G2 | F7 | 0.739127 | 4 | 0 | 4 |
| ATP6V1G2 | OLFM1 | 0.715973 | 3 | 0 | 3 |
| ATP6V1G2 | LSM11 | 0.708363 | 4 | 0 | 4 |
| ATP6V1G2 | NIPAL3 | 0.703865 | 3 | 0 | 3 |
| ATP6V1G2 | ZNF776 | 0.698057 | 3 | 0 | 3 |
| ATP6V1G2 | DNTT | 0.69555 | 4 | 0 | 4 |
| ATP6V1G2 | IQUB | 0.694052 | 3 | 0 | 3 |
| ATP6V1G2 | GBE1 | 0.689096 | 4 | 0 | 4 |
| ATP6V1G2 | DRD5 | 0.686301 | 4 | 0 | 4 |
| ATP6V1G2 | PURA | 0.68389 | 5 | 0 | 5 |
| ATP6V1G2 | BRK1 | 0.682817 | 3 | 0 | 3 |
| ATP6V1G2 | ZFAND5 | 0.681356 | 4 | 0 | 3 |
| ATP6V1G2 | GPRASP2 | 0.68051 | 3 | 0 | 3 |
| ATP6V1G2 | PLXNB1 | 0.679966 | 4 | 0 | 4 |
| ATP6V1G2 | NDUFC1 | 0.679889 | 3 | 0 | 3 |
| ATP6V1G2 | C5orf24 | 0.676485 | 4 | 0 | 4 |
For details and further investigation, click here