| Full name: galectin 13 | Alias Symbol: PP13|PLAC8 | ||
| Type: protein-coding gene | Cytoband: 19q13.2 | ||
| Entrez ID: 29124 | HGNC ID: HGNC:15449 | Ensembl Gene: ENSG00000105198 | OMIM ID: 608717 |
| Drug and gene relationship at DGIdb | |||
Expression of LGALS13:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | LGALS13 | 29124 | 220440_at | -0.1185 | 0.5716 | |
| GSE20347 | LGALS13 | 29124 | 220440_at | 0.0574 | 0.4023 | |
| GSE23400 | LGALS13 | 29124 | 220440_at | -0.1513 | 0.0031 | |
| GSE26886 | LGALS13 | 29124 | 220440_at | -0.1964 | 0.0631 | |
| GSE29001 | LGALS13 | 29124 | 220440_at | -0.0790 | 0.6948 | |
| GSE38129 | LGALS13 | 29124 | 220440_at | -0.0526 | 0.4213 | |
| GSE45670 | LGALS13 | 29124 | 220440_at | -0.0071 | 0.9468 | |
| GSE53622 | LGALS13 | 29124 | 73609 | -0.0327 | 0.7603 | |
| GSE53624 | LGALS13 | 29124 | 73609 | 0.0085 | 0.9219 | |
| GSE63941 | LGALS13 | 29124 | 220440_at | 0.1319 | 0.3566 | |
| GSE77861 | LGALS13 | 29124 | 220440_at | -0.1006 | 0.2824 |
Upregulated datasets: 0; Downregulated datasets: 0.
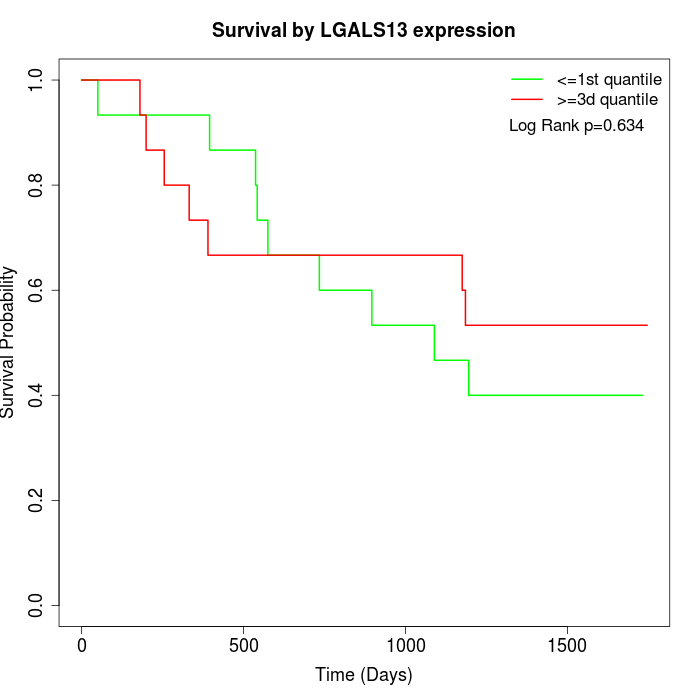
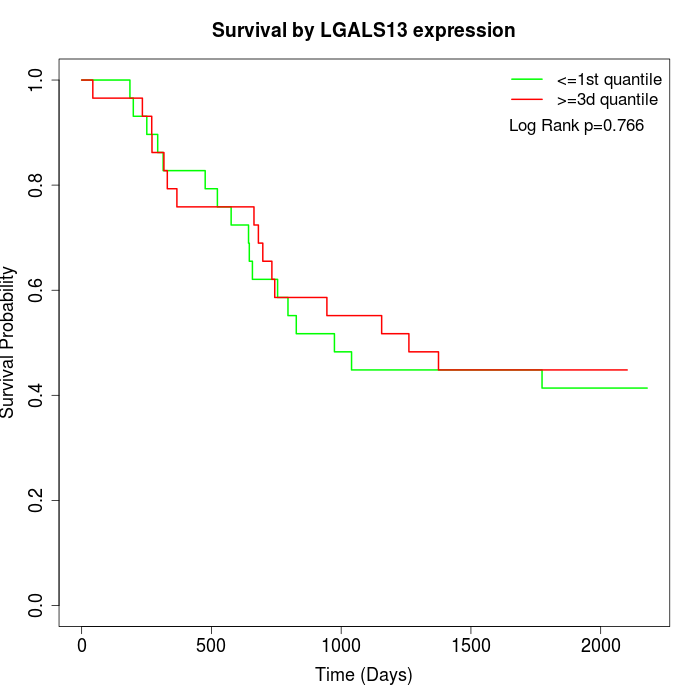
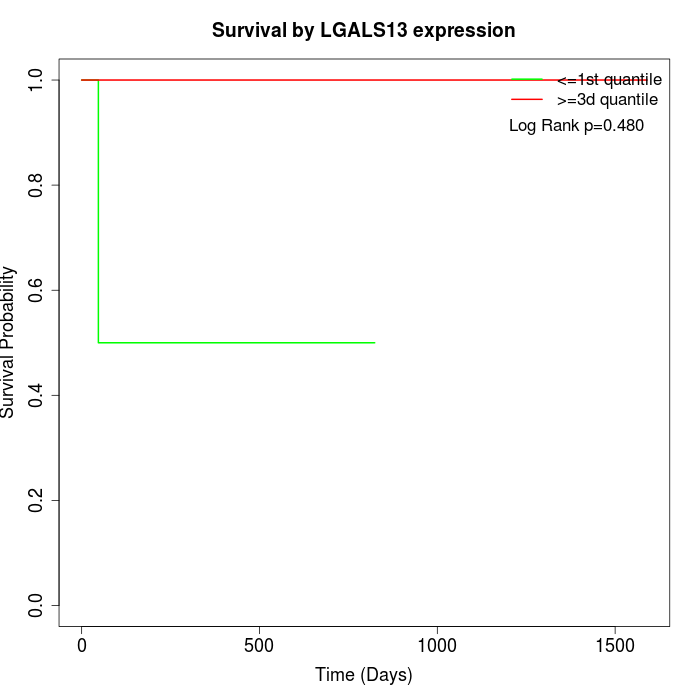
Survival by LGALS13 expression:
Note: Click image to view full size file.
Copy number change of LGALS13:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | LGALS13 | 29124 | 7 | 4 | 19 | |
| GSE20123 | LGALS13 | 29124 | 7 | 3 | 20 | |
| GSE43470 | LGALS13 | 29124 | 3 | 11 | 29 | |
| GSE46452 | LGALS13 | 29124 | 48 | 1 | 10 | |
| GSE47630 | LGALS13 | 29124 | 9 | 5 | 26 | |
| GSE54993 | LGALS13 | 29124 | 17 | 3 | 50 | |
| GSE54994 | LGALS13 | 29124 | 7 | 9 | 37 | |
| GSE60625 | LGALS13 | 29124 | 9 | 0 | 2 | |
| GSE74703 | LGALS13 | 29124 | 3 | 7 | 26 | |
| GSE74704 | LGALS13 | 29124 | 7 | 1 | 12 | |
| TCGA | LGALS13 | 29124 | 14 | 16 | 66 |
Total number of gains: 131; Total number of losses: 60; Total Number of normals: 297.
Somatic mutations of LGALS13:
Generating mutation plots.
Highly correlated genes for LGALS13:
Showing top 20/635 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| LGALS13 | BFSP2 | 0.757885 | 3 | 0 | 3 |
| LGALS13 | FGF14 | 0.751957 | 3 | 0 | 3 |
| LGALS13 | AMOT | 0.711186 | 4 | 0 | 4 |
| LGALS13 | FGF5 | 0.698447 | 3 | 0 | 3 |
| LGALS13 | CLDN9 | 0.696523 | 3 | 0 | 3 |
| LGALS13 | IFNA16 | 0.695379 | 4 | 0 | 4 |
| LGALS13 | HBD | 0.685947 | 3 | 0 | 3 |
| LGALS13 | RNF17 | 0.680961 | 4 | 0 | 4 |
| LGALS13 | STAC | 0.680561 | 3 | 0 | 3 |
| LGALS13 | AICDA | 0.677745 | 3 | 0 | 3 |
| LGALS13 | SLC5A4 | 0.677565 | 4 | 0 | 4 |
| LGALS13 | PRKG1 | 0.674923 | 4 | 0 | 3 |
| LGALS13 | CCDC30 | 0.673728 | 4 | 0 | 4 |
| LGALS13 | STXBP6 | 0.672838 | 4 | 0 | 3 |
| LGALS13 | IHH | 0.672183 | 3 | 0 | 3 |
| LGALS13 | CLCN4 | 0.667794 | 5 | 0 | 5 |
| LGALS13 | GDF10 | 0.665435 | 4 | 0 | 4 |
| LGALS13 | KRTAP19-3 | 0.665157 | 3 | 0 | 3 |
| LGALS13 | NRIP2 | 0.662448 | 3 | 0 | 3 |
| LGALS13 | RUNDC3A | 0.661929 | 3 | 0 | 3 |
For details and further investigation, click here