| Full name: B and T lymphocyte associated | Alias Symbol: BTLA1|CD272 | ||
| Type: protein-coding gene | Cytoband: 3q13.2 | ||
| Entrez ID: 151888 | HGNC ID: HGNC:21087 | Ensembl Gene: ENSG00000186265 | OMIM ID: 607925 |
| Drug and gene relationship at DGIdb | |||
Expression of BTLA:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BTLA | 151888 | 236226_at | 0.1234 | 0.8602 | |
| GSE26886 | BTLA | 151888 | 236226_at | -0.3158 | 0.0359 | |
| GSE45670 | BTLA | 151888 | 236226_at | 0.1123 | 0.3624 | |
| GSE53622 | BTLA | 151888 | 5563 | -0.8549 | 0.0032 | |
| GSE53624 | BTLA | 151888 | 5563 | -1.2583 | 0.0000 | |
| GSE63941 | BTLA | 151888 | 236226_at | 0.0328 | 0.8450 | |
| GSE77861 | BTLA | 151888 | 236226_at | -0.0851 | 0.5000 | |
| GSE97050 | BTLA | 151888 | A_33_P3358923 | 0.6057 | 0.3217 | |
| SRP133303 | BTLA | 151888 | RNAseq | -0.5083 | 0.1144 | |
| SRP219564 | BTLA | 151888 | RNAseq | 0.6588 | 0.3750 | |
| TCGA | BTLA | 151888 | RNAseq | -0.5988 | 0.1895 |
Upregulated datasets: 0; Downregulated datasets: 1.
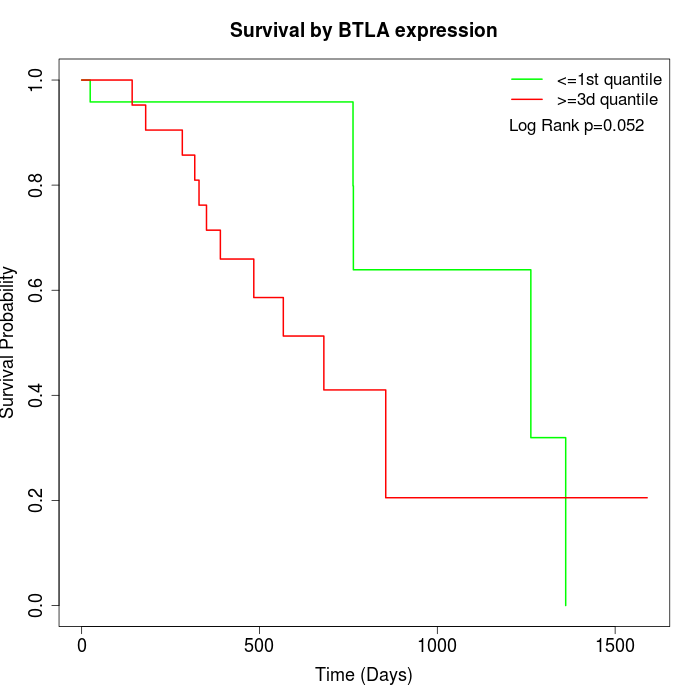
Survival by BTLA expression:
Note: Click image to view full size file.
Copy number change of BTLA:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BTLA | 151888 | 17 | 1 | 12 | |
| GSE20123 | BTLA | 151888 | 17 | 1 | 12 | |
| GSE43470 | BTLA | 151888 | 20 | 1 | 22 | |
| GSE46452 | BTLA | 151888 | 12 | 6 | 41 | |
| GSE47630 | BTLA | 151888 | 16 | 6 | 18 | |
| GSE54993 | BTLA | 151888 | 2 | 5 | 63 | |
| GSE54994 | BTLA | 151888 | 29 | 4 | 20 | |
| GSE60625 | BTLA | 151888 | 0 | 6 | 5 | |
| GSE74703 | BTLA | 151888 | 16 | 1 | 19 | |
| GSE74704 | BTLA | 151888 | 13 | 1 | 6 | |
| TCGA | BTLA | 151888 | 52 | 7 | 37 |
Total number of gains: 194; Total number of losses: 39; Total Number of normals: 255.
Somatic mutations of BTLA:
Generating mutation plots.
Highly correlated genes for BTLA:
Showing top 20/288 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BTLA | CD79B | 0.8586 | 4 | 0 | 4 |
| BTLA | KLRB1 | 0.810565 | 5 | 0 | 5 |
| BTLA | GPR18 | 0.772913 | 6 | 0 | 6 |
| BTLA | HLA-DOA | 0.77082 | 4 | 0 | 4 |
| BTLA | TRAT1 | 0.765335 | 5 | 0 | 5 |
| BTLA | CD48 | 0.764252 | 5 | 0 | 5 |
| BTLA | CD180 | 0.76406 | 5 | 0 | 5 |
| BTLA | IRF4 | 0.757975 | 6 | 0 | 6 |
| BTLA | PIM2 | 0.752558 | 3 | 0 | 3 |
| BTLA | LINC01215 | 0.749034 | 5 | 0 | 5 |
| BTLA | COL23A1 | 0.745774 | 3 | 0 | 3 |
| BTLA | CD37 | 0.745745 | 5 | 0 | 4 |
| BTLA | BANK1 | 0.745457 | 5 | 0 | 5 |
| BTLA | GRAP2 | 0.744272 | 3 | 0 | 3 |
| BTLA | CORO1A | 0.740074 | 6 | 0 | 6 |
| BTLA | GIMAP1 | 0.739067 | 5 | 0 | 5 |
| BTLA | ITK | 0.737032 | 6 | 0 | 5 |
| BTLA | ARHGAP15 | 0.735611 | 3 | 0 | 3 |
| BTLA | SNX20 | 0.734492 | 6 | 0 | 6 |
| BTLA | P2RY10 | 0.73261 | 6 | 0 | 5 |
For details and further investigation, click here