| Full name: T cell receptor associated transmembrane adaptor 1 | Alias Symbol: HSPC062|TRIM | ||
| Type: protein-coding gene | Cytoband: 3q13.13 | ||
| Entrez ID: 50852 | HGNC ID: HGNC:30698 | Ensembl Gene: ENSG00000163519 | OMIM ID: 604962 |
| Drug and gene relationship at DGIdb | |||
Expression of TRAT1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | TRAT1 | 50852 | 217147_s_at | 0.1992 | 0.8430 | |
| GSE20347 | TRAT1 | 50852 | 217147_s_at | -0.1059 | 0.0936 | |
| GSE23400 | TRAT1 | 50852 | 217147_s_at | -0.0299 | 0.1876 | |
| GSE26886 | TRAT1 | 50852 | 217147_s_at | -0.2834 | 0.0404 | |
| GSE29001 | TRAT1 | 50852 | 217147_s_at | -0.1442 | 0.3319 | |
| GSE38129 | TRAT1 | 50852 | 217147_s_at | -0.1713 | 0.1896 | |
| GSE45670 | TRAT1 | 50852 | 217147_s_at | -0.1790 | 0.4858 | |
| GSE53622 | TRAT1 | 50852 | 122816 | -0.8536 | 0.0002 | |
| GSE53624 | TRAT1 | 50852 | 122816 | -1.0930 | 0.0000 | |
| GSE63941 | TRAT1 | 50852 | 217147_s_at | -0.0364 | 0.8044 | |
| GSE77861 | TRAT1 | 50852 | 217147_s_at | -0.0418 | 0.6178 | |
| GSE97050 | TRAT1 | 50852 | A_23_P212568 | 0.7872 | 0.2569 | |
| SRP133303 | TRAT1 | 50852 | RNAseq | -0.6682 | 0.1624 | |
| TCGA | TRAT1 | 50852 | RNAseq | 0.2277 | 0.6723 |
Upregulated datasets: 0; Downregulated datasets: 1.
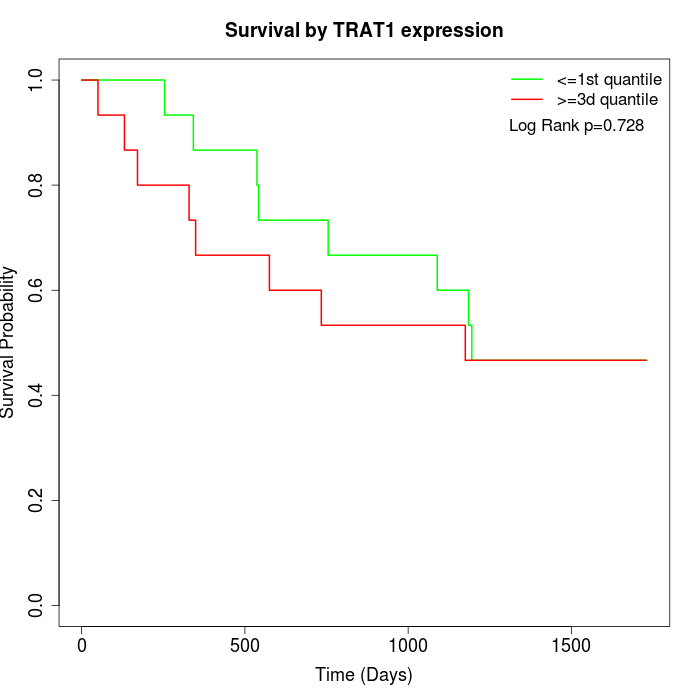
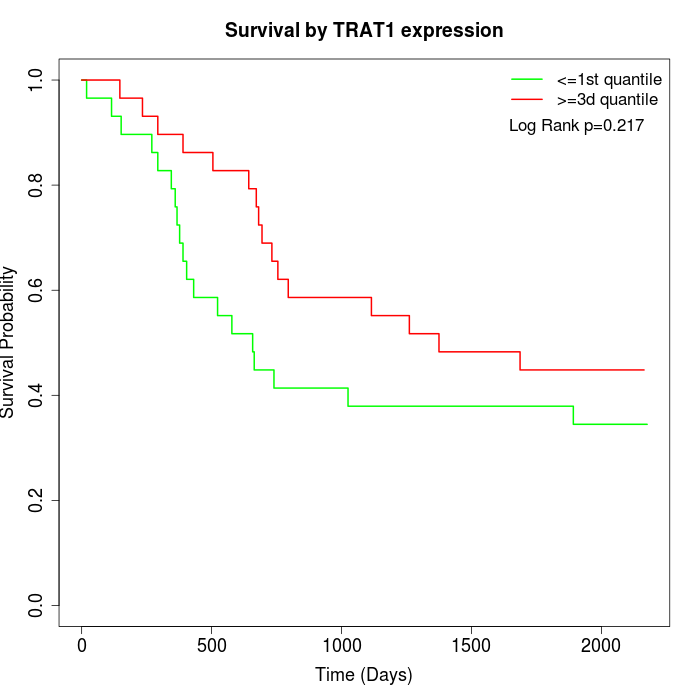
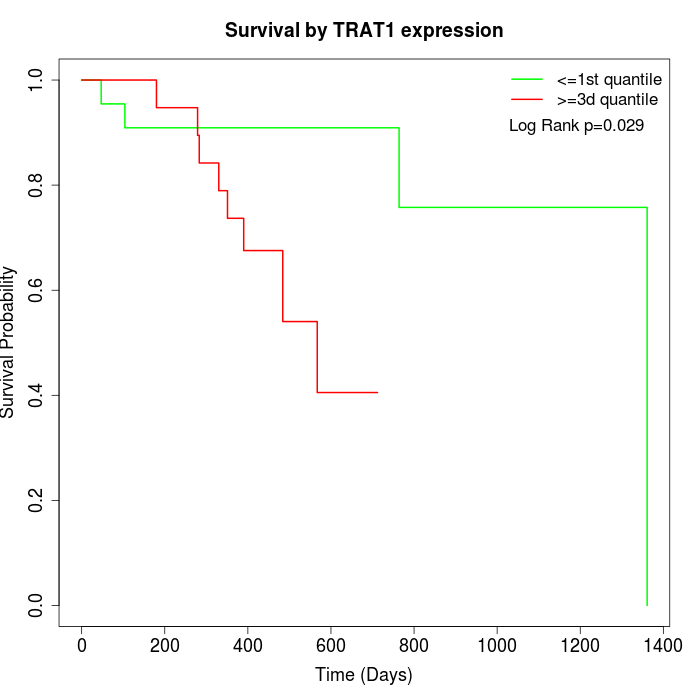
Survival by TRAT1 expression:
Note: Click image to view full size file.
Copy number change of TRAT1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | TRAT1 | 50852 | 17 | 1 | 12 | |
| GSE20123 | TRAT1 | 50852 | 16 | 1 | 13 | |
| GSE43470 | TRAT1 | 50852 | 18 | 1 | 24 | |
| GSE46452 | TRAT1 | 50852 | 12 | 6 | 41 | |
| GSE47630 | TRAT1 | 50852 | 16 | 6 | 18 | |
| GSE54993 | TRAT1 | 50852 | 2 | 5 | 63 | |
| GSE54994 | TRAT1 | 50852 | 28 | 5 | 20 | |
| GSE60625 | TRAT1 | 50852 | 0 | 6 | 5 | |
| GSE74703 | TRAT1 | 50852 | 15 | 1 | 20 | |
| GSE74704 | TRAT1 | 50852 | 13 | 1 | 6 | |
| TCGA | TRAT1 | 50852 | 50 | 8 | 38 |
Total number of gains: 187; Total number of losses: 41; Total Number of normals: 260.
Somatic mutations of TRAT1:
Generating mutation plots.
Highly correlated genes for TRAT1:
Showing top 20/307 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| TRAT1 | FAM78A | 0.782039 | 3 | 0 | 3 |
| TRAT1 | BTLA | 0.765335 | 5 | 0 | 5 |
| TRAT1 | GNGT2 | 0.764987 | 3 | 0 | 3 |
| TRAT1 | NLRC3 | 0.761739 | 5 | 0 | 5 |
| TRAT1 | TAGAP | 0.757512 | 4 | 0 | 4 |
| TRAT1 | CD48 | 0.756586 | 9 | 0 | 8 |
| TRAT1 | TRAF3IP3 | 0.743016 | 8 | 0 | 7 |
| TRAT1 | AKNA | 0.7321 | 4 | 0 | 4 |
| TRAT1 | THEMIS | 0.725261 | 5 | 0 | 4 |
| TRAT1 | DTHD1 | 0.718977 | 3 | 0 | 3 |
| TRAT1 | PLCG2 | 0.714725 | 3 | 0 | 3 |
| TRAT1 | ITK | 0.70627 | 9 | 0 | 7 |
| TRAT1 | GIMAP7 | 0.702619 | 5 | 0 | 5 |
| TRAT1 | BLK | 0.699186 | 4 | 0 | 4 |
| TRAT1 | FGD3 | 0.69678 | 3 | 0 | 3 |
| TRAT1 | SASH3 | 0.693628 | 10 | 0 | 8 |
| TRAT1 | TIGIT | 0.693493 | 5 | 0 | 4 |
| TRAT1 | GPR18 | 0.69258 | 9 | 0 | 7 |
| TRAT1 | CD3D | 0.692082 | 10 | 0 | 8 |
| TRAT1 | CD2 | 0.689722 | 10 | 0 | 8 |
For details and further investigation, click here