| Full name: beta-transducin repeat containing E3 ubiquitin protein ligase | Alias Symbol: bTrCP|betaTrCP|FBXW1A|Fwd1|beta-TrCP1|bTrCP1 | ||
| Type: protein-coding gene | Cytoband: 10q24.32 | ||
| Entrez ID: 8945 | HGNC ID: HGNC:1144 | Ensembl Gene: ENSG00000166167 | OMIM ID: 603482 |
| Drug and gene relationship at DGIdb | |||
Screen Evidence:
| |||
BTRC involved pathways:
| KEGG pathway | Description | View |
|---|---|---|
| hsa04114 | Oocyte meiosis | |
| hsa04310 | Wnt signaling pathway | |
| hsa04340 | Hedgehog signaling pathway | |
| hsa04390 | Hippo signaling pathway | |
| hsa04710 | Circadian rhythm | |
| hsa05131 | Shigellosis |
Expression of BTRC:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | BTRC | 8945 | 224471_s_at | -0.1899 | 0.6661 | |
| GSE20347 | BTRC | 8945 | 204901_at | -0.2962 | 0.0212 | |
| GSE23400 | BTRC | 8945 | 204901_at | -0.1960 | 0.0000 | |
| GSE26886 | BTRC | 8945 | 224471_s_at | -0.4721 | 0.0445 | |
| GSE29001 | BTRC | 8945 | 204901_at | -0.6765 | 0.0203 | |
| GSE38129 | BTRC | 8945 | 204901_at | -0.2685 | 0.0173 | |
| GSE45670 | BTRC | 8945 | 224471_s_at | -0.0820 | 0.5690 | |
| GSE53622 | BTRC | 8945 | 21499 | -0.3977 | 0.0000 | |
| GSE53624 | BTRC | 8945 | 21499 | -0.4509 | 0.0000 | |
| GSE63941 | BTRC | 8945 | 224471_s_at | -0.9034 | 0.0117 | |
| GSE77861 | BTRC | 8945 | 224471_s_at | -0.1627 | 0.3886 | |
| GSE97050 | BTRC | 8945 | A_23_P46819 | 0.0313 | 0.9132 | |
| SRP007169 | BTRC | 8945 | RNAseq | -1.2288 | 0.0027 | |
| SRP008496 | BTRC | 8945 | RNAseq | -0.9249 | 0.0001 | |
| SRP064894 | BTRC | 8945 | RNAseq | -0.6691 | 0.0000 | |
| SRP133303 | BTRC | 8945 | RNAseq | -0.2723 | 0.0145 | |
| SRP159526 | BTRC | 8945 | RNAseq | -0.5981 | 0.1073 | |
| SRP193095 | BTRC | 8945 | RNAseq | -0.2625 | 0.0028 | |
| SRP219564 | BTRC | 8945 | RNAseq | -0.4363 | 0.0776 | |
| TCGA | BTRC | 8945 | RNAseq | -0.1144 | 0.0287 |
Upregulated datasets: 0; Downregulated datasets: 1.
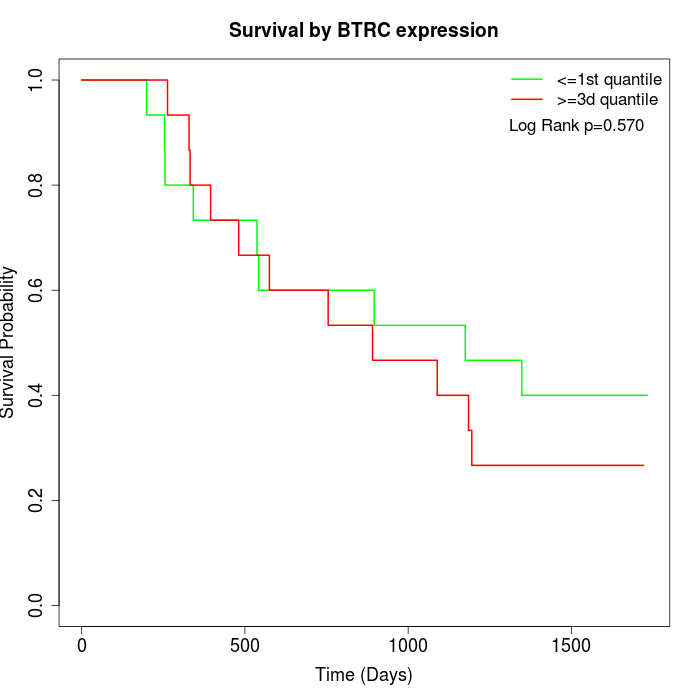
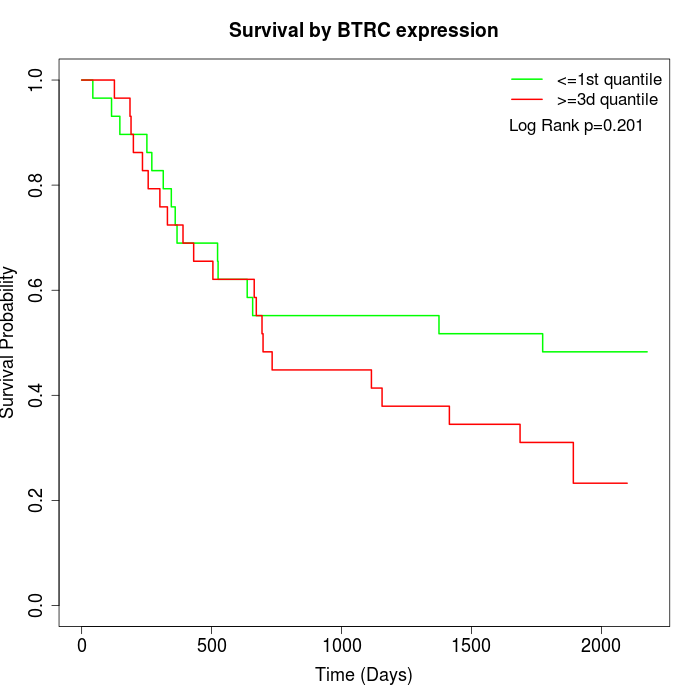
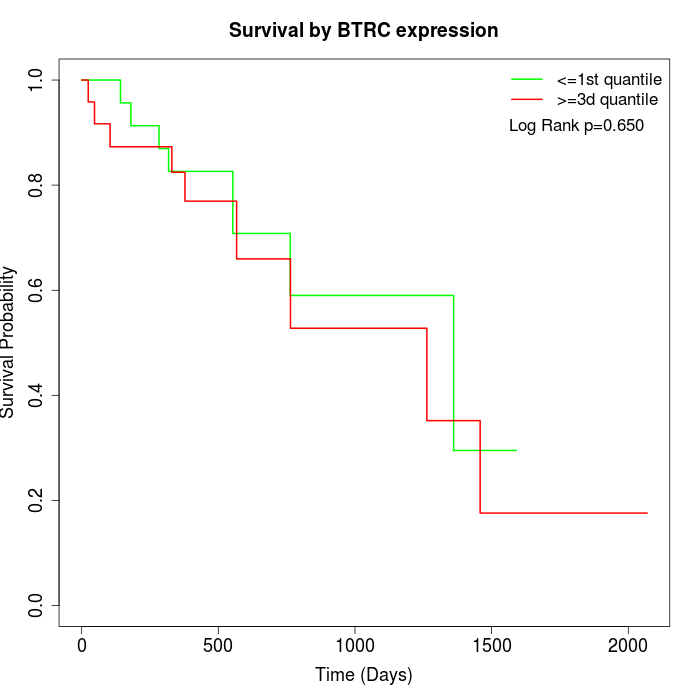
Survival by BTRC expression:
Note: Click image to view full size file.
Copy number change of BTRC:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | BTRC | 8945 | 2 | 9 | 19 | |
| GSE20123 | BTRC | 8945 | 2 | 8 | 20 | |
| GSE43470 | BTRC | 8945 | 0 | 8 | 35 | |
| GSE46452 | BTRC | 8945 | 0 | 11 | 48 | |
| GSE47630 | BTRC | 8945 | 2 | 14 | 24 | |
| GSE54993 | BTRC | 8945 | 8 | 0 | 62 | |
| GSE54994 | BTRC | 8945 | 1 | 13 | 39 | |
| GSE60625 | BTRC | 8945 | 0 | 0 | 11 | |
| GSE74703 | BTRC | 8945 | 0 | 5 | 31 | |
| GSE74704 | BTRC | 8945 | 1 | 4 | 15 | |
| TCGA | BTRC | 8945 | 5 | 28 | 63 |
Total number of gains: 21; Total number of losses: 100; Total Number of normals: 367.
Somatic mutations of BTRC:
Generating mutation plots.
Highly correlated genes for BTRC:
Showing top 20/718 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| BTRC | SYPL1 | 0.809142 | 3 | 0 | 3 |
| BTRC | TRAPPC6B | 0.7842 | 3 | 0 | 3 |
| BTRC | RHBDD1 | 0.748636 | 3 | 0 | 3 |
| BTRC | TMEM126A | 0.74672 | 3 | 0 | 3 |
| BTRC | GLYR1 | 0.746184 | 3 | 0 | 3 |
| BTRC | NANS | 0.738834 | 3 | 0 | 3 |
| BTRC | DIS3L | 0.737468 | 3 | 0 | 3 |
| BTRC | ZNF564 | 0.731515 | 3 | 0 | 3 |
| BTRC | ZNF814 | 0.72922 | 3 | 0 | 3 |
| BTRC | ZRANB1 | 0.723982 | 4 | 0 | 4 |
| BTRC | CCIN | 0.722648 | 5 | 0 | 5 |
| BTRC | FAM160B1 | 0.712487 | 3 | 0 | 3 |
| BTRC | ZSCAN29 | 0.70738 | 3 | 0 | 3 |
| BTRC | DUSP28 | 0.705797 | 3 | 0 | 3 |
| BTRC | TMTC3 | 0.700143 | 3 | 0 | 3 |
| BTRC | FBXW8 | 0.693796 | 3 | 0 | 3 |
| BTRC | GNAT2 | 0.689828 | 3 | 0 | 3 |
| BTRC | IFT74 | 0.689645 | 3 | 0 | 3 |
| BTRC | BDH2 | 0.686753 | 3 | 0 | 3 |
| BTRC | PDCD7 | 0.68657 | 4 | 0 | 4 |
For details and further investigation, click here