| Full name: C1QTNF1 antisense RNA 1 | Alias Symbol: | ||
| Type: non-coding RNA | Cytoband: 17q25.3 | ||
| Entrez ID: 100507410 | HGNC ID: HGNC:44351 | Ensembl Gene: ENSG00000265096 | OMIM ID: |
| Drug and gene relationship at DGIdb | |||
Expression of C1QTNF1-AS1:
| Dataset | Gene | EntrezID | Probe | Log2FC | Adj.pValue | Expression |
|---|---|---|---|---|---|---|
| GSE17351 | C1QTNF1-AS1 | 100507410 | 237313_at | 0.0003 | 0.9993 | |
| GSE26886 | C1QTNF1-AS1 | 100507410 | 237313_at | 0.1104 | 0.3004 | |
| GSE45670 | C1QTNF1-AS1 | 100507410 | 237313_at | -0.0904 | 0.2666 | |
| GSE53622 | C1QTNF1-AS1 | 100507410 | 1729 | 0.1669 | 0.2334 | |
| GSE53624 | C1QTNF1-AS1 | 100507410 | 1729 | 0.6213 | 0.0000 | |
| GSE63941 | C1QTNF1-AS1 | 100507410 | 237313_at | -0.1123 | 0.5743 | |
| GSE77861 | C1QTNF1-AS1 | 100507410 | 237313_at | -0.0784 | 0.3886 |
Upregulated datasets: 0; Downregulated datasets: 0.
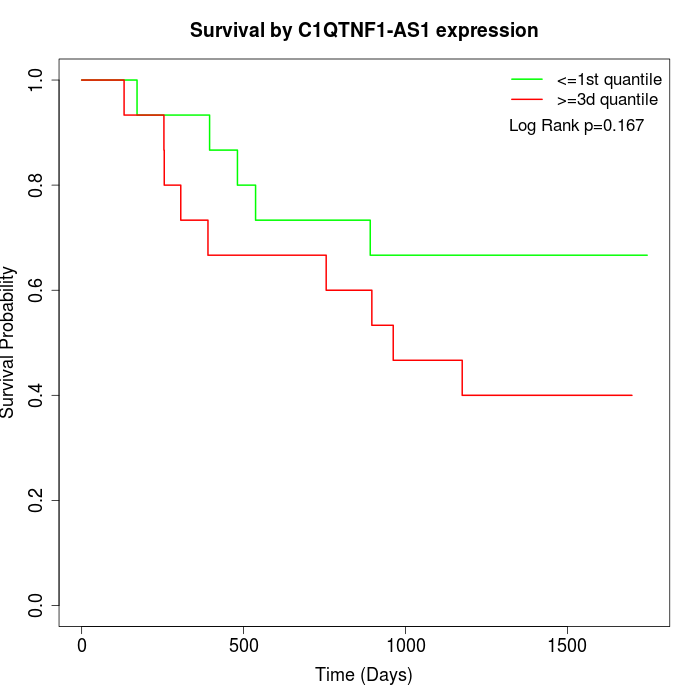
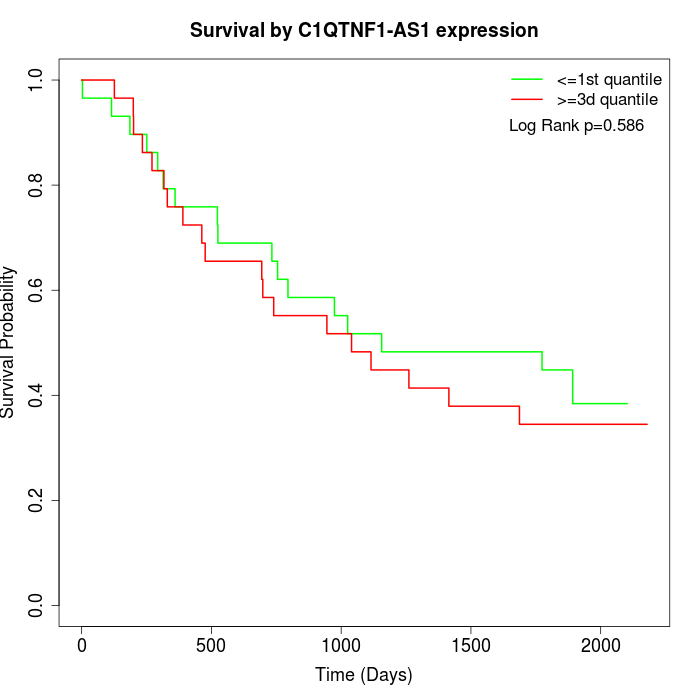
Survival by C1QTNF1-AS1 expression:
Note: Click image to view full size file.
Copy number change of C1QTNF1-AS1:
| Dataset | Gene | EntrezID | Gain | Loss | Normal | Detail |
|---|---|---|---|---|---|---|
| GSE15526 | C1QTNF1-AS1 | 100507410 | 4 | 3 | 23 | |
| GSE20123 | C1QTNF1-AS1 | 100507410 | 4 | 3 | 23 | |
| GSE43470 | C1QTNF1-AS1 | 100507410 | 3 | 4 | 36 | |
| GSE46452 | C1QTNF1-AS1 | 100507410 | 33 | 0 | 26 | |
| GSE47630 | C1QTNF1-AS1 | 100507410 | 8 | 1 | 31 | |
| GSE54993 | C1QTNF1-AS1 | 100507410 | 2 | 5 | 63 | |
| GSE54994 | C1QTNF1-AS1 | 100507410 | 10 | 5 | 38 | |
| GSE60625 | C1QTNF1-AS1 | 100507410 | 6 | 0 | 5 | |
| GSE74703 | C1QTNF1-AS1 | 100507410 | 3 | 2 | 31 | |
| GSE74704 | C1QTNF1-AS1 | 100507410 | 4 | 3 | 13 |
Total number of gains: 77; Total number of losses: 26; Total Number of normals: 289.
Somatic mutations of C1QTNF1-AS1:
Generating mutation plots.
Highly correlated genes for C1QTNF1-AS1:
Showing top 20/43 corelated genes with mean PCC>0.5.
| Gene1 | Gene2 | Mean PCC | Num. Datasets | Num. PCC<0 | Num. PCC>0.5 |
|---|---|---|---|---|---|
| C1QTNF1-AS1 | CYP2W1 | 0.706178 | 3 | 0 | 3 |
| C1QTNF1-AS1 | SSTR3 | 0.69444 | 3 | 0 | 3 |
| C1QTNF1-AS1 | OR3A2 | 0.667799 | 3 | 0 | 3 |
| C1QTNF1-AS1 | DRD4 | 0.663074 | 3 | 0 | 3 |
| C1QTNF1-AS1 | EVX1 | 0.655738 | 3 | 0 | 3 |
| C1QTNF1-AS1 | VMO1 | 0.651662 | 3 | 0 | 3 |
| C1QTNF1-AS1 | DEFA4 | 0.649459 | 3 | 0 | 3 |
| C1QTNF1-AS1 | PYDC1 | 0.633906 | 3 | 0 | 3 |
| C1QTNF1-AS1 | SNX8 | 0.6304 | 3 | 0 | 3 |
| C1QTNF1-AS1 | NPTXR | 0.627536 | 3 | 0 | 3 |
| C1QTNF1-AS1 | KCNQ1DN | 0.62368 | 3 | 0 | 3 |
| C1QTNF1-AS1 | APOA4 | 0.623277 | 3 | 0 | 3 |
| C1QTNF1-AS1 | NKAIN4 | 0.618262 | 3 | 0 | 3 |
| C1QTNF1-AS1 | INHBE | 0.616179 | 3 | 0 | 3 |
| C1QTNF1-AS1 | PDE3A | 0.615858 | 3 | 0 | 3 |
| C1QTNF1-AS1 | LINC01341 | 0.612092 | 3 | 0 | 3 |
| C1QTNF1-AS1 | CFC1B | 0.607059 | 3 | 0 | 3 |
| C1QTNF1-AS1 | CLCN2 | 0.600248 | 4 | 0 | 3 |
| C1QTNF1-AS1 | TH | 0.598007 | 4 | 0 | 3 |
| C1QTNF1-AS1 | CLDN9 | 0.59465 | 5 | 0 | 3 |
For details and further investigation, click here